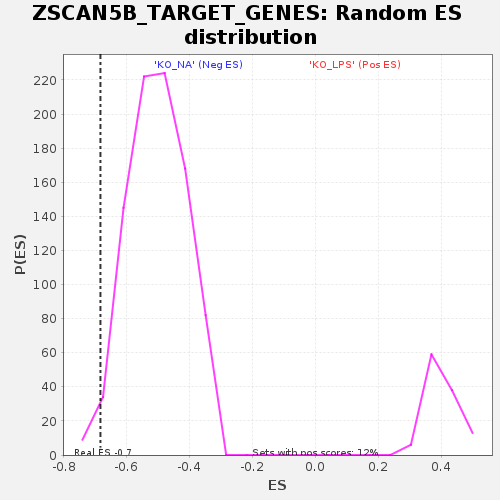
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | ZSCAN5B_TARGET_GENES |
| Enrichment Score (ES) | -0.68267405 |
| Normalized Enrichment Score (NES) | -1.3589183 |
| Nominal p-value | 0.021493213 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | LASP1 | LIM and SH3 protein 1 [Source:HGNC Symbol;Acc:HGNC:6513] | 1262 | 1.339 | -0.0114 | No |
| 2 | BCAR3 | BCAR3 adaptor protein, NSP family member [Source:HGNC Symbol;Acc:HGNC:973] | 1409 | 1.242 | 0.0057 | No |
| 3 | POLR3F | RNA polymerase III subunit F [Source:HGNC Symbol;Acc:HGNC:15763] | 2088 | 0.903 | 0.0027 | No |
| 4 | RAB11A | RAB11A, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9760] | 2521 | 0.734 | 0.0035 | No |
| 5 | MYNN | myoneurin [Source:HGNC Symbol;Acc:HGNC:14955] | 3146 | 0.565 | -0.0037 | No |
| 6 | HSPA6 | heat shock protein family A (Hsp70) member 6 [Source:HGNC Symbol;Acc:HGNC:5239] | 3505 | 0.479 | -0.0053 | No |
| 7 | PSMB3 | proteasome 20S subunit beta 3 [Source:HGNC Symbol;Acc:HGNC:9540] | 3517 | 0.476 | 0.0025 | No |
| 8 | DPP9 | dipeptidyl peptidase 9 [Source:HGNC Symbol;Acc:HGNC:18648] | 3620 | 0.454 | 0.0074 | No |
| 9 | TRIM41 | tripartite motif containing 41 [Source:HGNC Symbol;Acc:HGNC:19013] | 3924 | 0.394 | 0.0059 | No |
| 10 | PRMT5 | protein arginine methyltransferase 5 [Source:HGNC Symbol;Acc:HGNC:10894] | 4018 | 0.370 | 0.0097 | No |
| 11 | ILF2 | interleukin enhancer binding factor 2 [Source:HGNC Symbol;Acc:HGNC:6037] | 5852 | 0.021 | -0.0395 | No |
| 12 | LINC01623 | long intergenic non-protein coding RNA 1623 [Source:HGNC Symbol;Acc:HGNC:52050] | 5967 | 0.001 | -0.0425 | No |
| 13 | PPEF1 | protein phosphatase with EF-hand domain 1 [Source:HGNC Symbol;Acc:HGNC:9243] | 6651 | 0.000 | -0.0610 | No |
| 14 | RNASE11 | ribonuclease A family member 11 (inactive) [Source:HGNC Symbol;Acc:HGNC:19269] | 7271 | 0.000 | -0.0777 | No |
| 15 | LINC01556 | long intergenic non-protein coding RNA 1556 [Source:HGNC Symbol;Acc:HGNC:21195] | 7460 | 0.000 | -0.0828 | No |
| 16 | H4C8 | H4 clustered histone 8 [Source:HGNC Symbol;Acc:HGNC:4788] | 9900 | 0.000 | -0.1487 | No |
| 17 | GSTA4 | glutathione S-transferase alpha 4 [Source:HGNC Symbol;Acc:HGNC:4629] | 9944 | 0.000 | -0.1499 | No |
| 18 | VTRNA1-2 | vault RNA 1-2 [Source:HGNC Symbol;Acc:HGNC:12655] | 10239 | 0.000 | -0.1578 | No |
| 19 | VTRNA1-1 | vault RNA 1-1 [Source:HGNC Symbol;Acc:HGNC:12654] | 10241 | 0.000 | -0.1578 | No |
| 20 | VTRNA1-3 | vault RNA 1-3 [Source:HGNC Symbol;Acc:HGNC:12656] | 10246 | 0.000 | -0.1579 | No |
| 21 | VTRNA2-1 | vault RNA 2-1 [Source:HGNC Symbol;Acc:HGNC:37054] | 10294 | 0.000 | -0.1592 | No |
| 22 | RNU6-1 | RNA, U6 small nuclear 1 [Source:HGNC Symbol;Acc:HGNC:10227] | 10564 | 0.000 | -0.1665 | No |
| 23 | RNU6-2 | RNA, U6 small nuclear 2 [Source:HGNC Symbol;Acc:HGNC:34270] | 10565 | 0.000 | -0.1665 | No |
| 24 | RNU6-9 | RNA, U6 small nuclear 9 [Source:HGNC Symbol;Acc:HGNC:34269] | 10569 | 0.000 | -0.1666 | No |
| 25 | AP001107.9 | novel transcript, antisense to SLC29A2 | 11435 | 0.000 | -0.1899 | No |
| 26 | GHET1 | gastric carcinoma proliferation enhancing transcript 1 [Source:HGNC Symbol;Acc:HGNC:49425] | 12787 | 0.000 | -0.2264 | No |
| 27 | AL132780.1 | novel transcript, antisense to HAUS4 | 13699 | 0.000 | -0.2510 | No |
| 28 | LINC02136 | long intergenic non-protein coding RNA 2136 [Source:HGNC Symbol;Acc:HGNC:52995] | 14113 | 0.000 | -0.2622 | No |
| 29 | STK17A | serine/threonine kinase 17a [Source:HGNC Symbol;Acc:HGNC:11395] | 14552 | 0.000 | -0.2740 | No |
| 30 | LINC02453 | long intergenic non-protein coding RNA 2453 [Source:HGNC Symbol;Acc:HGNC:53392] | 14740 | 0.000 | -0.2791 | No |
| 31 | LIMD1-AS1 | LIMD1 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:44107] | 16366 | 0.000 | -0.3230 | No |
| 32 | HJV | hemojuvelin BMP co-receptor [Source:HGNC Symbol;Acc:HGNC:4887] | 17356 | 0.000 | -0.3497 | No |
| 33 | AGBL5-AS1 | AGBL5 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:41133] | 17656 | 0.000 | -0.3578 | No |
| 34 | HCG14 | HLA complex group 14 [Source:HGNC Symbol;Acc:HGNC:18323] | 18444 | 0.000 | -0.3791 | No |
| 35 | RAD51AP2 | RAD51 associated protein 2 [Source:HGNC Symbol;Acc:HGNC:34417] | 20529 | 0.000 | -0.4354 | No |
| 36 | MAP1LC3B | microtubule associated protein 1 light chain 3 beta [Source:HGNC Symbol;Acc:HGNC:13352] | 22170 | 0.000 | -0.4797 | No |
| 37 | RNY4 | RNA, Ro60-associated Y4 [Source:HGNC Symbol;Acc:HGNC:10244] | 22537 | 0.000 | -0.4896 | No |
| 38 | RNY1 | RNA, Ro60-associated Y1 [Source:HGNC Symbol;Acc:HGNC:10242] | 22538 | 0.000 | -0.4896 | No |
| 39 | HMGN4 | high mobility group nucleosomal binding domain 4 [Source:HGNC Symbol;Acc:HGNC:4989] | 24391 | 0.000 | -0.5396 | No |
| 40 | RNU6ATAC | RNA, U6atac small nuclear (U12-dependent splicing) [Source:HGNC Symbol;Acc:HGNC:34017] | 25435 | 0.000 | -0.5678 | No |
| 41 | RN7SK | RNA component of 7SK nuclear ribonucleoprotein [Source:HGNC Symbol;Acc:HGNC:10037] | 27683 | -0.058 | -0.6275 | No |
| 42 | ZNF839 | zinc finger protein 839 [Source:HGNC Symbol;Acc:HGNC:20345] | 28110 | -0.121 | -0.6370 | No |
| 43 | FMC1-LUC7L2 | FMC1-LUC7L2 readthrough [Source:HGNC Symbol;Acc:HGNC:44671] | 28121 | -0.122 | -0.6352 | No |
| 44 | STAP2 | signal transducing adaptor family member 2 [Source:HGNC Symbol;Acc:HGNC:30430] | 29192 | -0.279 | -0.6593 | No |
| 45 | POLG | DNA polymerase gamma, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9179] | 29205 | -0.282 | -0.6549 | No |
| 46 | ALOXE3 | arachidonate lipoxygenase 3 [Source:HGNC Symbol;Acc:HGNC:13743] | 29451 | -0.319 | -0.6561 | No |
| 47 | DZANK1 | double zinc ribbon and ankyrin repeat domains 1 [Source:HGNC Symbol;Acc:HGNC:15858] | 30437 | -0.465 | -0.6748 | Yes |
| 48 | PPP6R3 | protein phosphatase 6 regulatory subunit 3 [Source:HGNC Symbol;Acc:HGNC:1173] | 30448 | -0.466 | -0.6671 | Yes |
| 49 | H3C9P | H3 clustered histone 9, pseudogene [Source:HGNC Symbol;Acc:HGNC:18982] | 30645 | -0.502 | -0.6639 | Yes |
| 50 | LRP3 | LDL receptor related protein 3 [Source:HGNC Symbol;Acc:HGNC:6695] | 30742 | -0.516 | -0.6578 | Yes |
| 51 | SLC27A4 | solute carrier family 27 member 4 [Source:HGNC Symbol;Acc:HGNC:10998] | 31191 | -0.594 | -0.6598 | Yes |
| 52 | POLR3E | RNA polymerase III subunit E [Source:HGNC Symbol;Acc:HGNC:30347] | 31513 | -0.654 | -0.6574 | Yes |
| 53 | NCOA7 | nuclear receptor coactivator 7 [Source:HGNC Symbol;Acc:HGNC:21081] | 31873 | -0.724 | -0.6548 | Yes |
| 54 | TMEM259 | transmembrane protein 259 [Source:HGNC Symbol;Acc:HGNC:17039] | 32170 | -0.801 | -0.6492 | Yes |
| 55 | GARNL3 | GTPase activating Rap/RanGAP domain like 3 [Source:HGNC Symbol;Acc:HGNC:25425] | 32669 | -0.935 | -0.6468 | Yes |
| 56 | RPS29 | ribosomal protein S29 [Source:HGNC Symbol;Acc:HGNC:10419] | 32861 | -0.993 | -0.6351 | Yes |
| 57 | SACM1L | SAC1 like phosphatidylinositide phosphatase [Source:HGNC Symbol;Acc:HGNC:17059] | 32888 | -1.002 | -0.6188 | Yes |
| 58 | SHF | Src homology 2 domain containing F [Source:HGNC Symbol;Acc:HGNC:25116] | 33189 | -1.092 | -0.6084 | Yes |
| 59 | SMARCC2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily c member 2 [Source:HGNC Symbol;Acc:HGNC:11105] | 33265 | -1.117 | -0.5914 | Yes |
| 60 | NDC1 | NDC1 transmembrane nucleoporin [Source:HGNC Symbol;Acc:HGNC:25525] | 33279 | -1.121 | -0.5728 | Yes |
| 61 | MED16 | mediator complex subunit 16 [Source:HGNC Symbol;Acc:HGNC:17556] | 33403 | -1.161 | -0.5564 | Yes |
| 62 | PARP2 | poly(ADP-ribose) polymerase 2 [Source:HGNC Symbol;Acc:HGNC:272] | 33694 | -1.264 | -0.5428 | Yes |
| 63 | SYPL1 | synaptophysin like 1 [Source:HGNC Symbol;Acc:HGNC:11507] | 33851 | -1.319 | -0.5246 | Yes |
| 64 | MCRIP2 | MAPK regulated corepressor interacting protein 2 [Source:HGNC Symbol;Acc:HGNC:14142] | 34161 | -1.432 | -0.5087 | Yes |
| 65 | CCDC107 | coiled-coil domain containing 107 [Source:HGNC Symbol;Acc:HGNC:28465] | 34376 | -1.516 | -0.4887 | Yes |
| 66 | FMC1 | formation of mitochondrial complex V assembly factor 1 homolog [Source:HGNC Symbol;Acc:HGNC:26946] | 34449 | -1.545 | -0.4645 | Yes |
| 67 | LTA4H | leukotriene A4 hydrolase [Source:HGNC Symbol;Acc:HGNC:6710] | 34765 | -1.683 | -0.4444 | Yes |
| 68 | NDUFS7 | NADH:ubiquinone oxidoreductase core subunit S7 [Source:HGNC Symbol;Acc:HGNC:7714] | 34804 | -1.700 | -0.4166 | Yes |
| 69 | TIA1 | TIA1 cytotoxic granule associated RNA binding protein [Source:HGNC Symbol;Acc:HGNC:11802] | 34911 | -1.758 | -0.3897 | Yes |
| 70 | FBXO31 | F-box protein 31 [Source:HGNC Symbol;Acc:HGNC:16510] | 35328 | -2.012 | -0.3668 | Yes |
| 71 | C2orf42 | chromosome 2 open reading frame 42 [Source:HGNC Symbol;Acc:HGNC:26056] | 35384 | -2.051 | -0.3334 | Yes |
| 72 | ANG | angiogenin [Source:HGNC Symbol;Acc:HGNC:483] | 35625 | -2.216 | -0.3023 | Yes |
| 73 | B4GAT1 | beta-1,4-glucuronyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:15685] | 35657 | -2.240 | -0.2652 | Yes |
| 74 | PLXDC1 | plexin domain containing 1 [Source:HGNC Symbol;Acc:HGNC:20945] | 35834 | -2.405 | -0.2291 | Yes |
| 75 | C8orf37 | chromosome 8 open reading frame 37 [Source:HGNC Symbol;Acc:HGNC:27232] | 36257 | -2.799 | -0.1930 | Yes |
| 76 | KNL1 | kinetochore scaffold 1 [Source:HGNC Symbol;Acc:HGNC:24054] | 36396 | -2.980 | -0.1462 | Yes |
| 77 | METTL26 | methyltransferase like 26 [Source:HGNC Symbol;Acc:HGNC:14141] | 36432 | -3.026 | -0.0958 | Yes |
| 78 | RNASE4 | ribonuclease A family member 4 [Source:HGNC Symbol;Acc:HGNC:10047] | 36455 | -3.057 | -0.0445 | Yes |
| 79 | AGBL5 | ATP/GTP binding protein like 5 [Source:HGNC Symbol;Acc:HGNC:26147] | 36804 | -3.634 | 0.0077 | Yes |