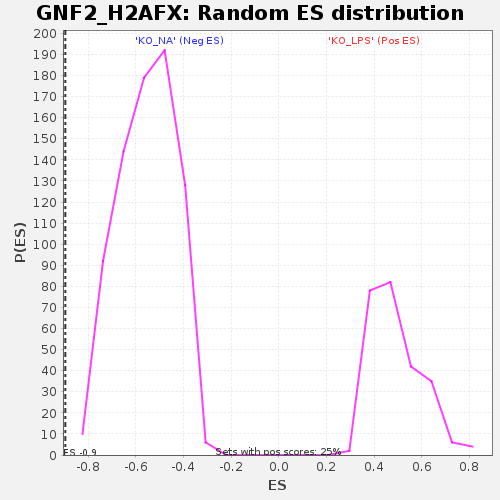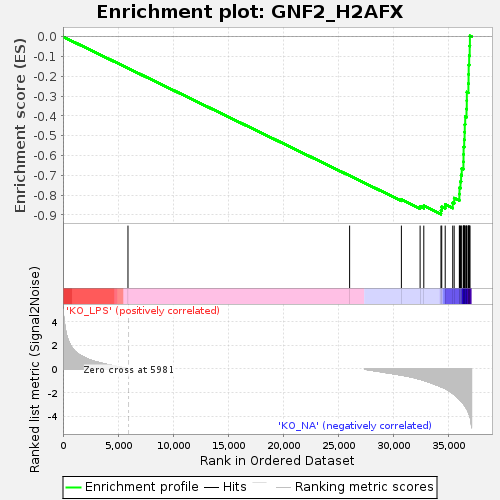
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_H2AFX |
| Enrichment Score (ES) | -0.89563966 |
| Normalized Enrichment Score (NES) | -1.6183321 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.001005276 |
| FWER p-Value | 0.022 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | UBE2S | ubiquitin conjugating enzyme E2 S [Source:HGNC Symbol;Acc:HGNC:17895] | 5898 | 0.011 | -0.1590 | No |
| 2 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26019 | 0.000 | -0.7019 | No |
| 3 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 30719 | -0.514 | -0.8221 | No |
| 4 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32418 | -0.869 | -0.8568 | No |
| 5 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 32753 | -0.958 | -0.8536 | No |
| 6 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 34312 | -1.491 | -0.8766 | Yes |
| 7 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 34381 | -1.518 | -0.8590 | Yes |
| 8 | NSD2 | nuclear receptor binding SET domain protein 2 [Source:HGNC Symbol;Acc:HGNC:12766] | 34697 | -1.649 | -0.8464 | Yes |
| 9 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 35380 | -2.050 | -0.8386 | Yes |
| 10 | DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 [Source:HGNC Symbol;Acc:HGNC:19123] | 35510 | -2.132 | -0.8148 | Yes |
| 11 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 35981 | -2.530 | -0.7952 | Yes |
| 12 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 35993 | -2.546 | -0.7629 | Yes |
| 13 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 36083 | -2.637 | -0.7316 | Yes |
| 14 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 36138 | -2.694 | -0.6986 | Yes |
| 15 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 36199 | -2.753 | -0.6650 | Yes |
| 16 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36350 | -2.926 | -0.6317 | Yes |
| 17 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 36359 | -2.931 | -0.5944 | Yes |
| 18 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36367 | -2.940 | -0.5570 | Yes |
| 19 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36421 | -3.011 | -0.5199 | Yes |
| 20 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36457 | -3.057 | -0.4818 | Yes |
| 21 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 36473 | -3.076 | -0.4429 | Yes |
| 22 | CCNF | cyclin F [Source:HGNC Symbol;Acc:HGNC:1591] | 36511 | -3.130 | -0.4039 | Yes |
| 23 | TMPO | thymopoietin [Source:HGNC Symbol;Acc:HGNC:11875] | 36629 | -3.321 | -0.3645 | Yes |
| 24 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 36655 | -3.354 | -0.3223 | Yes |
| 25 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36660 | -3.369 | -0.2794 | Yes |
| 26 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36802 | -3.632 | -0.2367 | Yes |
| 27 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 36817 | -3.672 | -0.1902 | Yes |
| 28 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36828 | -3.703 | -0.1431 | Yes |
| 29 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 36873 | -3.821 | -0.0954 | Yes |
| 30 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36920 | -3.941 | -0.0463 | Yes |
| 31 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36939 | -3.976 | 0.0041 | Yes |