Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
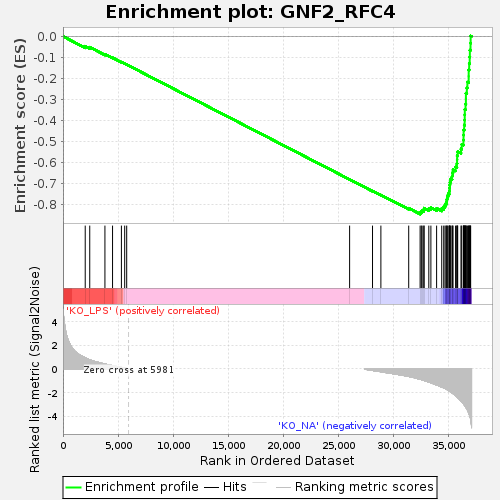
| GeneSet | GNF2_RFC4 |
| Enrichment Score (ES) | -0.8452428 |
| Normalized Enrichment Score (NES) | -1.6451226 |
| Nominal p-value | 0.0 |
| FDR q-value | 4.0841842E-4 |
| FWER p-Value | 0.007 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
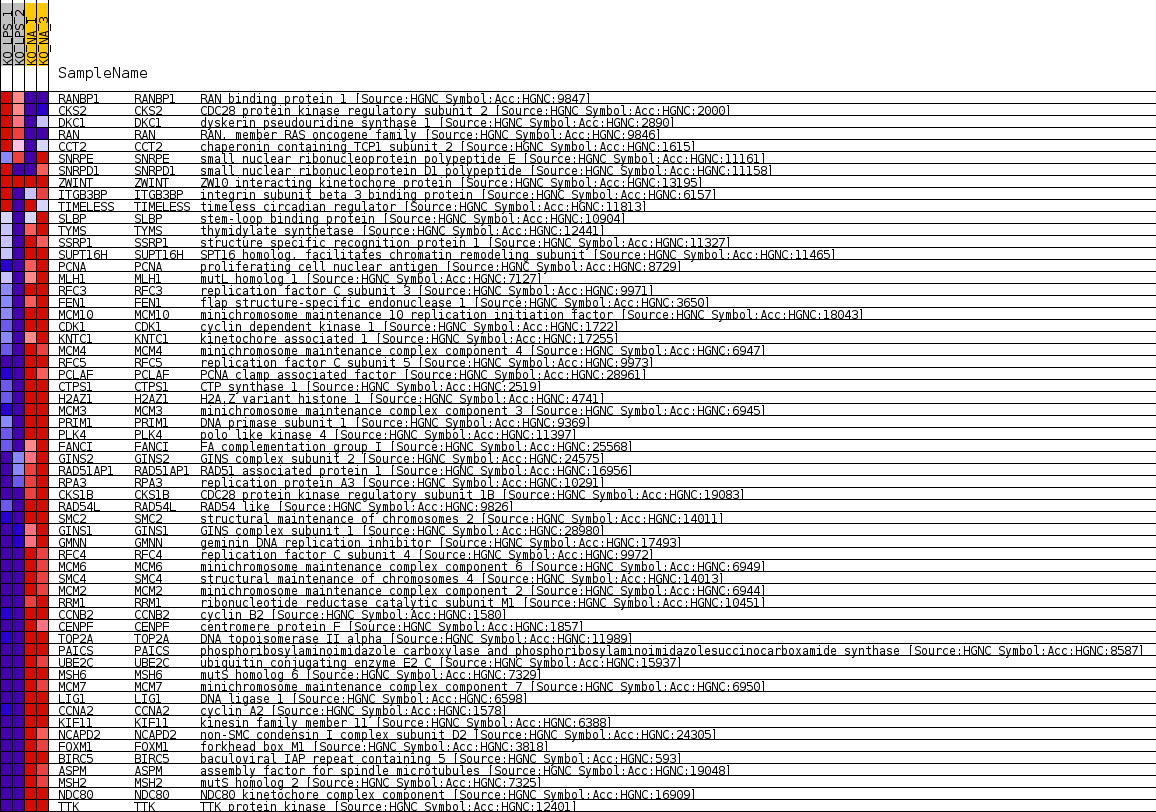
| 1 | RANBP1 | RAN binding protein 1 [Source:HGNC Symbol;Acc:HGNC:9847] | 2017 | 0.936 | -0.0467 | No |
| 2 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 2428 | 0.770 | -0.0515 | No |
| 3 | DKC1 | dyskerin pseudouridine synthase 1 [Source:HGNC Symbol;Acc:HGNC:2890] | 3806 | 0.420 | -0.0852 | No |
| 4 | RAN | RAN, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9846] | 4498 | 0.278 | -0.1015 | No |
| 5 | CCT2 | chaperonin containing TCP1 subunit 2 [Source:HGNC Symbol;Acc:HGNC:1615] | 5302 | 0.130 | -0.1221 | No |
| 6 | SNRPE | small nuclear ribonucleoprotein polypeptide E [Source:HGNC Symbol;Acc:HGNC:11161] | 5600 | 0.072 | -0.1296 | No |
| 7 | SNRPD1 | small nuclear ribonucleoprotein D1 polypeptide [Source:HGNC Symbol;Acc:HGNC:11158] | 5782 | 0.036 | -0.1342 | No |
| 8 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26019 | 0.000 | -0.6806 | No |
| 9 | ITGB3BP | integrin subunit beta 3 binding protein [Source:HGNC Symbol;Acc:HGNC:6157] | 28098 | -0.120 | -0.7357 | No |
| 10 | TIMELESS | timeless circadian regulator [Source:HGNC Symbol;Acc:HGNC:11813] | 28857 | -0.228 | -0.7543 | No |
| 11 | SLBP | stem-loop binding protein [Source:HGNC Symbol;Acc:HGNC:10904] | 31383 | -0.630 | -0.8173 | No |
| 12 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32418 | -0.869 | -0.8381 | Yes |
| 13 | SSRP1 | structure specific recognition protein 1 [Source:HGNC Symbol;Acc:HGNC:11327] | 32525 | -0.892 | -0.8336 | Yes |
| 14 | SUPT16H | SPT16 homolog, facilitates chromatin remodeling subunit [Source:HGNC Symbol;Acc:HGNC:11465] | 32601 | -0.917 | -0.8280 | Yes |
| 15 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 32753 | -0.958 | -0.8242 | Yes |
| 16 | MLH1 | mutL homolog 1 [Source:HGNC Symbol;Acc:HGNC:7127] | 32781 | -0.966 | -0.8169 | Yes |
| 17 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33208 | -1.098 | -0.8194 | Yes |
| 18 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 33397 | -1.158 | -0.8149 | Yes |
| 19 | MCM10 | minichromosome maintenance 10 replication initiation factor [Source:HGNC Symbol;Acc:HGNC:18043] | 33916 | -1.342 | -0.8178 | Yes |
| 20 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 34381 | -1.518 | -0.8178 | Yes |
| 21 | KNTC1 | kinetochore associated 1 [Source:HGNC Symbol;Acc:HGNC:17255] | 34562 | -1.584 | -0.8095 | Yes |
| 22 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34684 | -1.643 | -0.7992 | Yes |
| 23 | RFC5 | replication factor C subunit 5 [Source:HGNC Symbol;Acc:HGNC:9973] | 34783 | -1.690 | -0.7879 | Yes |
| 24 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 34837 | -1.712 | -0.7752 | Yes |
| 25 | CTPS1 | CTP synthase 1 [Source:HGNC Symbol;Acc:HGNC:2519] | 34882 | -1.740 | -0.7620 | Yes |
| 26 | H2AZ1 | H2A.Z variant histone 1 [Source:HGNC Symbol;Acc:HGNC:4741] | 34958 | -1.777 | -0.7494 | Yes |
| 27 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 35062 | -1.840 | -0.7370 | Yes |
| 28 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35085 | -1.852 | -0.7223 | Yes |
| 29 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 35101 | -1.865 | -0.7073 | Yes |
| 30 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35131 | -1.888 | -0.6925 | Yes |
| 31 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35192 | -1.927 | -0.6782 | Yes |
| 32 | RAD51AP1 | RAD51 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16956] | 35318 | -2.003 | -0.6650 | Yes |
| 33 | RPA3 | replication protein A3 [Source:HGNC Symbol;Acc:HGNC:10291] | 35348 | -2.023 | -0.6491 | Yes |
| 34 | CKS1B | CDC28 protein kinase regulatory subunit 1B [Source:HGNC Symbol;Acc:HGNC:19083] | 35407 | -2.067 | -0.6336 | Yes |
| 35 | RAD54L | RAD54 like [Source:HGNC Symbol;Acc:HGNC:9826] | 35638 | -2.227 | -0.6214 | Yes |
| 36 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35759 | -2.331 | -0.6054 | Yes |
| 37 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 35776 | -2.350 | -0.5864 | Yes |
| 38 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 35779 | -2.351 | -0.5670 | Yes |
| 39 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 35830 | -2.402 | -0.5485 | Yes |
| 40 | MCM6 | minichromosome maintenance complex component 6 [Source:HGNC Symbol;Acc:HGNC:6949] | 36129 | -2.683 | -0.5344 | Yes |
| 41 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 36199 | -2.753 | -0.5135 | Yes |
| 42 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36350 | -2.926 | -0.4934 | Yes |
| 43 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 36359 | -2.931 | -0.4694 | Yes |
| 44 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36367 | -2.940 | -0.4453 | Yes |
| 45 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36421 | -3.011 | -0.4219 | Yes |
| 46 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 36461 | -3.062 | -0.3977 | Yes |
| 47 | PAICS | phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase [Source:HGNC Symbol;Acc:HGNC:8587] | 36469 | -3.069 | -0.3725 | Yes |
| 48 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 36476 | -3.083 | -0.3472 | Yes |
| 49 | MSH6 | mutS homolog 6 [Source:HGNC Symbol;Acc:HGNC:7329] | 36542 | -3.181 | -0.3227 | Yes |
| 50 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36575 | -3.228 | -0.2969 | Yes |
| 51 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36576 | -3.229 | -0.2702 | Yes |
| 52 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36660 | -3.369 | -0.2446 | Yes |
| 53 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 36709 | -3.440 | -0.2175 | Yes |
| 54 | NCAPD2 | non-SMC condensin I complex subunit D2 [Source:HGNC Symbol;Acc:HGNC:24305] | 36827 | -3.702 | -0.1901 | Yes |
| 55 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36828 | -3.703 | -0.1595 | Yes |
| 56 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 36873 | -3.821 | -0.1291 | Yes |
| 57 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36920 | -3.941 | -0.0978 | Yes |
| 58 | MSH2 | mutS homolog 2 [Source:HGNC Symbol;Acc:HGNC:7325] | 36935 | -3.972 | -0.0654 | Yes |
| 59 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36994 | -4.180 | -0.0324 | Yes |
| 60 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 37006 | -4.236 | 0.0023 | Yes |