Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_LPS |
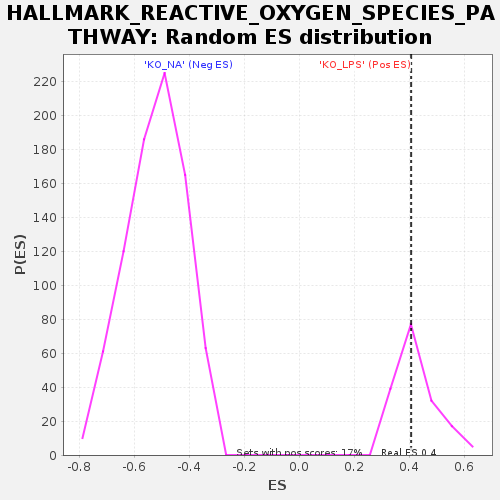
| GeneSet | HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY |
| Enrichment Score (ES) | 0.40735534 |
| Normalized Enrichment Score (NES) | 0.9529153 |
| Nominal p-value | 0.5176471 |
| FDR q-value | 0.5832923 |
| FWER p-Value | 0.923 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | JUNB | JunB proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:6205] | 156 | 3.689 | 0.0607 | Yes |
| 2 | SOD2 | superoxide dismutase 2 [Source:HGNC Symbol;Acc:HGNC:11180] | 216 | 3.338 | 0.1178 | Yes |
| 3 | GLRX | glutaredoxin [Source:HGNC Symbol;Acc:HGNC:4330] | 218 | 3.326 | 0.1764 | Yes |
| 4 | TXNRD1 | thioredoxin reductase 1 [Source:HGNC Symbol;Acc:HGNC:12437] | 340 | 2.796 | 0.2223 | Yes |
| 5 | SRXN1 | sulfiredoxin 1 [Source:HGNC Symbol;Acc:HGNC:16132] | 502 | 2.362 | 0.2595 | Yes |
| 6 | PRDX1 | peroxiredoxin 1 [Source:HGNC Symbol;Acc:HGNC:9352] | 795 | 1.824 | 0.2837 | Yes |
| 7 | GCLM | glutamate-cysteine ligase modifier subunit [Source:HGNC Symbol;Acc:HGNC:4312] | 818 | 1.790 | 0.3146 | Yes |
| 8 | SELENOS | selenoprotein S [Source:HGNC Symbol;Acc:HGNC:30396] | 835 | 1.775 | 0.3454 | Yes |
| 9 | TXN | thioredoxin [Source:HGNC Symbol;Acc:HGNC:12435] | 985 | 1.591 | 0.3694 | Yes |
| 10 | ABCC1 | ATP binding cassette subfamily C member 1 [Source:HGNC Symbol;Acc:HGNC:51] | 1071 | 1.498 | 0.3934 | Yes |
| 11 | SBNO2 | strawberry notch homolog 2 [Source:HGNC Symbol;Acc:HGNC:29158] | 1675 | 1.104 | 0.3966 | Yes |
| 12 | EGLN2 | egl-9 family hypoxia inducible factor 2 [Source:HGNC Symbol;Acc:HGNC:14660] | 1920 | 0.987 | 0.4074 | Yes |
| 13 | SCAF4 | SR-related CTD associated factor 4 [Source:HGNC Symbol;Acc:HGNC:19304] | 2432 | 0.767 | 0.4071 | No |
| 14 | LAMTOR5 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 [Source:HGNC Symbol;Acc:HGNC:17955] | 2857 | 0.635 | 0.4068 | No |
| 15 | G6PD | glucose-6-phosphate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4057] | 3352 | 0.517 | 0.4025 | No |
| 16 | NQO1 | NAD(P)H quinone dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:2874] | 4082 | 0.361 | 0.3892 | No |
| 17 | GSR | glutathione-disulfide reductase [Source:HGNC Symbol;Acc:HGNC:4623] | 4811 | 0.217 | 0.3734 | No |
| 18 | PRDX2 | peroxiredoxin 2 [Source:HGNC Symbol;Acc:HGNC:9353] | 5021 | 0.174 | 0.3708 | No |
| 19 | MSRA | methionine sulfoxide reductase A [Source:HGNC Symbol;Acc:HGNC:7377] | 5059 | 0.168 | 0.3728 | No |
| 20 | GPX3 | glutathione peroxidase 3 [Source:HGNC Symbol;Acc:HGNC:4555] | 9198 | 0.000 | 0.2611 | No |
| 21 | MGST1 | microsomal glutathione S-transferase 1 [Source:HGNC Symbol;Acc:HGNC:7061] | 11564 | 0.000 | 0.1972 | No |
| 22 | IPCEF1 | interaction protein for cytohesin exchange factors 1 [Source:HGNC Symbol;Acc:HGNC:21204] | 17864 | 0.000 | 0.0272 | No |
| 23 | MPO | myeloperoxidase [Source:HGNC Symbol;Acc:HGNC:7218] | 18953 | 0.000 | -0.0022 | No |
| 24 | NDUFB4 | NADH:ubiquinone oxidoreductase subunit B4 [Source:HGNC Symbol;Acc:HGNC:7699] | 22814 | 0.000 | -0.1064 | No |
| 25 | HHEX | hematopoietically expressed homeobox [Source:HGNC Symbol;Acc:HGNC:4901] | 28711 | -0.206 | -0.2620 | No |
| 26 | OXSR1 | oxidative stress responsive kinase 1 [Source:HGNC Symbol;Acc:HGNC:8508] | 28887 | -0.232 | -0.2626 | No |
| 27 | STK25 | serine/threonine kinase 25 [Source:HGNC Symbol;Acc:HGNC:11404] | 28938 | -0.240 | -0.2598 | No |
| 28 | FTL | ferritin light chain [Source:HGNC Symbol;Acc:HGNC:3999] | 29143 | -0.271 | -0.2605 | No |
| 29 | ATOX1 | antioxidant 1 copper chaperone [Source:HGNC Symbol;Acc:HGNC:798] | 29211 | -0.283 | -0.2573 | No |
| 30 | PRDX4 | peroxiredoxin 4 [Source:HGNC Symbol;Acc:HGNC:17169] | 29510 | -0.329 | -0.2596 | No |
| 31 | PDLIM1 | PDZ and LIM domain 1 [Source:HGNC Symbol;Acc:HGNC:2067] | 29549 | -0.333 | -0.2547 | No |
| 32 | SOD1 | superoxide dismutase 1 [Source:HGNC Symbol;Acc:HGNC:11179] | 30037 | -0.403 | -0.2608 | No |
| 33 | ERCC2 | ERCC excision repair 2, TFIIH core complex helicase subunit [Source:HGNC Symbol;Acc:HGNC:3434] | 30096 | -0.414 | -0.2551 | No |
| 34 | PRNP | prion protein [Source:HGNC Symbol;Acc:HGNC:9449] | 30356 | -0.453 | -0.2541 | No |
| 35 | HMOX2 | heme oxygenase 2 [Source:HGNC Symbol;Acc:HGNC:5014] | 30553 | -0.486 | -0.2508 | No |
| 36 | TXNRD2 | thioredoxin reductase 2 [Source:HGNC Symbol;Acc:HGNC:18155] | 31044 | -0.567 | -0.2541 | No |
| 37 | GLRX2 | glutaredoxin 2 [Source:HGNC Symbol;Acc:HGNC:16065] | 32369 | -0.856 | -0.2748 | No |
| 38 | NDUFS2 | NADH:ubiquinone oxidoreductase core subunit S2 [Source:HGNC Symbol;Acc:HGNC:7708] | 33225 | -1.104 | -0.2784 | No |
| 39 | FES | FES proto-oncogene, tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:3657] | 33556 | -1.213 | -0.2660 | No |
| 40 | CAT | catalase [Source:HGNC Symbol;Acc:HGNC:1516] | 33693 | -1.263 | -0.2475 | No |
| 41 | NDUFA6 | NADH:ubiquinone oxidoreductase subunit A6 [Source:HGNC Symbol;Acc:HGNC:7690] | 33725 | -1.273 | -0.2259 | No |
| 42 | GCLC | glutamate-cysteine ligase catalytic subunit [Source:HGNC Symbol;Acc:HGNC:4311] | 33998 | -1.371 | -0.2091 | No |
| 43 | PTPA | protein phosphatase 2 phosphatase activator [Source:HGNC Symbol;Acc:HGNC:9308] | 34181 | -1.441 | -0.1887 | No |
| 44 | PRDX6 | peroxiredoxin 6 [Source:HGNC Symbol;Acc:HGNC:16753] | 34417 | -1.533 | -0.1680 | No |
| 45 | PFKP | phosphofructokinase, platelet [Source:HGNC Symbol;Acc:HGNC:8878] | 34466 | -1.552 | -0.1420 | No |
| 46 | LSP1 | lymphocyte specific protein 1 [Source:HGNC Symbol;Acc:HGNC:6707] | 35044 | -1.831 | -0.1254 | No |
| 47 | GPX4 | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 36617 | -3.295 | -0.1098 | No |
| 48 | CDKN2D | cyclin dependent kinase inhibitor 2D [Source:HGNC Symbol;Acc:HGNC:1790] | 36696 | -3.419 | -0.0518 | No |
| 49 | MBP | myelin basic protein [Source:HGNC Symbol;Acc:HGNC:6925] | 36767 | -3.545 | 0.0087 | No |