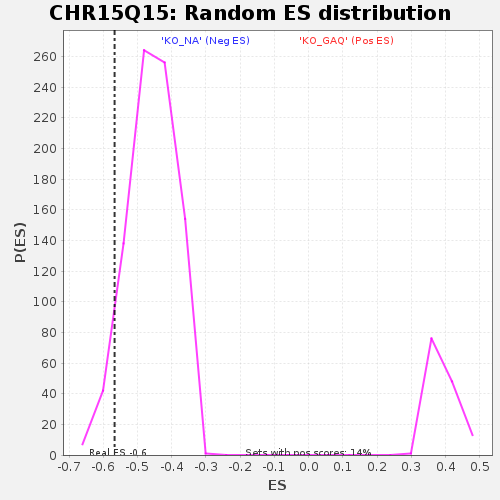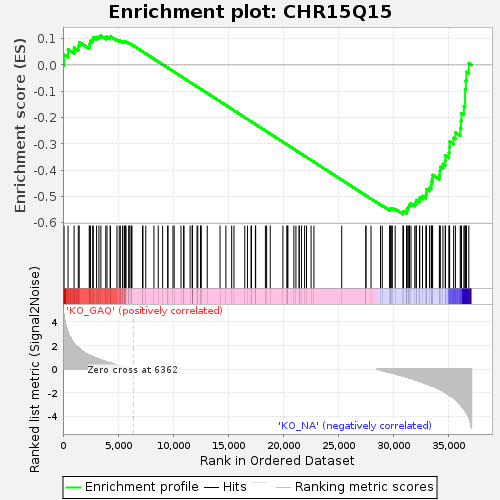
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | CHR15Q15 |
| Enrichment Score (ES) | -0.5669051 |
| Normalized Enrichment Score (NES) | -1.238992 |
| Nominal p-value | 0.061484918 |
| FDR q-value | 0.8324457 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | EXD1 | exonuclease 3'-5' domain containing 1 [Source:HGNC Symbol;Acc:HGNC:28507] | 96 | 4.402 | 0.0396 | No |
| 2 | ACTBP7 | ACTB pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:140] | 457 | 3.063 | 0.0593 | No |
| 3 | TMEM62 | transmembrane protein 62 [Source:HGNC Symbol;Acc:HGNC:26269] | 1005 | 2.137 | 0.0650 | No |
| 4 | MYL12BP1 | myosin light chain 12B pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:51649] | 1389 | 1.817 | 0.0720 | No |
| 5 | CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransferase 1 [Source:HGNC Symbol;Acc:HGNC:28680] | 1470 | 1.727 | 0.0864 | No |
| 6 | NDUFAF1 | NADH:ubiquinone oxidoreductase complex assembly factor 1 [Source:HGNC Symbol;Acc:HGNC:18828] | 2384 | 1.174 | 0.0730 | No |
| 7 | TMEM87A | transmembrane protein 87A [Source:HGNC Symbol;Acc:HGNC:24522] | 2440 | 1.153 | 0.0826 | No |
| 8 | MFAP1 | microfibril associated protein 1 [Source:HGNC Symbol;Acc:HGNC:7032] | 2490 | 1.132 | 0.0921 | No |
| 9 | MGA | MAX dimerization protein MGA [Source:HGNC Symbol;Acc:HGNC:14010] | 2695 | 1.050 | 0.0967 | No |
| 10 | ELOCP2 | elongin C pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:17762] | 2748 | 1.027 | 0.1051 | No |
| 11 | TP53BP1 | tumor protein p53 binding protein 1 [Source:HGNC Symbol;Acc:HGNC:11999] | 3048 | 0.906 | 0.1057 | No |
| 12 | RPUSD2 | RNA pseudouridine synthase domain containing 2 [Source:HGNC Symbol;Acc:HGNC:24180] | 3260 | 0.820 | 0.1079 | No |
| 13 | VPS18 | VPS18 core subunit of CORVET and HOPS complexes [Source:HGNC Symbol;Acc:HGNC:15972] | 3438 | 0.759 | 0.1104 | No |
| 14 | UBR1 | ubiquitin protein ligase E3 component n-recognin 1 [Source:HGNC Symbol;Acc:HGNC:16808] | 3882 | 0.612 | 0.1042 | No |
| 15 | SPCS2P1 | signal peptidase complex subunit 2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:31885] | 3995 | 0.581 | 0.1068 | No |
| 16 | INO80 | INO80 complex ATPase subunit [Source:HGNC Symbol;Acc:HGNC:26956] | 4276 | 0.496 | 0.1040 | No |
| 17 | LRRC57 | leucine rich repeat containing 57 [Source:HGNC Symbol;Acc:HGNC:26719] | 4310 | 0.486 | 0.1077 | No |
| 18 | SRP14 | signal recognition particle 14 [Source:HGNC Symbol;Acc:HGNC:11299] | 4899 | 0.318 | 0.0949 | No |
| 19 | CATSPER2 | cation channel sperm associated 2 [Source:HGNC Symbol;Acc:HGNC:18810] | 5103 | 0.256 | 0.0918 | No |
| 20 | RHOV | ras homolog family member V [Source:HGNC Symbol;Acc:HGNC:18313] | 5203 | 0.239 | 0.0914 | No |
| 21 | CYCSP2 | CYCS pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:24410] | 5404 | 0.239 | 0.0883 | No |
| 22 | ZSCAN29 | zinc finger and SCAN domain containing 29 [Source:HGNC Symbol;Acc:HGNC:26673] | 5557 | 0.234 | 0.0865 | No |
| 23 | BAHD1 | bromo adjacent homology domain containing 1 [Source:HGNC Symbol;Acc:HGNC:29153] | 5595 | 0.227 | 0.0876 | No |
| 24 | RMDN3 | regulator of microtubule dynamics 3 [Source:HGNC Symbol;Acc:HGNC:25550] | 5666 | 0.199 | 0.0877 | No |
| 25 | PIN4P1 | peptidylprolyl cis/trans isomerase, NIMA-interacting 4 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:44193] | 5723 | 0.185 | 0.0879 | No |
| 26 | HYPK | huntingtin interacting protein K [Source:HGNC Symbol;Acc:HGNC:18418] | 5979 | 0.105 | 0.0820 | No |
| 27 | LCMT2 | leucine carboxyl methyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:17558] | 5983 | 0.104 | 0.0829 | No |
| 28 | RTF1 | RTF1 homolog, Paf1/RNA polymerase II complex component [Source:HGNC Symbol;Acc:HGNC:28996] | 6084 | 0.073 | 0.0809 | No |
| 29 | SERF2 | small EDRK-rich factor 2 [Source:HGNC Symbol;Acc:HGNC:10757] | 6226 | 0.037 | 0.0775 | No |
| 30 | PDIA3 | protein disulfide isomerase family A member 3 [Source:HGNC Symbol;Acc:HGNC:4606] | 6257 | 0.026 | 0.0769 | No |
| 31 | C15orf62 | chromosome 15 open reading frame 62 [Source:HGNC Symbol;Acc:HGNC:34489] | 7221 | 0.000 | 0.0508 | No |
| 32 | C15orf56 | chromosome 15 open reading frame 56 [Source:HGNC Symbol;Acc:HGNC:33868] | 7258 | 0.000 | 0.0499 | No |
| 33 | RNU6-188P | RNA, U6 small nuclear 188, pseudogene [Source:HGNC Symbol;Acc:HGNC:47151] | 7515 | 0.000 | 0.0429 | No |
| 34 | EHD4-AS1 | EHD4 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:51418] | 8254 | 0.000 | 0.0230 | No |
| 35 | FRMD5 | FERM domain containing 5 [Source:HGNC Symbol;Acc:HGNC:28214] | 8646 | 0.000 | 0.0124 | No |
| 36 | SERINC4 | serine incorporator 4 [Source:HGNC Symbol;Acc:HGNC:32237] | 9045 | 0.000 | 0.0016 | No |
| 37 | AC016134.1 | novel transcript | 9502 | 0.000 | -0.0107 | No |
| 38 | RNU6-1169P | RNA, U6 small nuclear 1169, pseudogene [Source:HGNC Symbol;Acc:HGNC:48132] | 9532 | 0.000 | -0.0115 | No |
| 39 | RNU6-516P | RNA, U6 small nuclear 516, pseudogene [Source:HGNC Symbol;Acc:HGNC:47479] | 9980 | 0.000 | -0.0236 | No |
| 40 | ELL3 | elongation factor for RNA polymerase II 3 [Source:HGNC Symbol;Acc:HGNC:23113] | 10094 | 0.000 | -0.0267 | No |
| 41 | RNU6-554P | RNA, U6 small nuclear 554, pseudogene [Source:HGNC Symbol;Acc:HGNC:47517] | 10714 | 0.000 | -0.0434 | No |
| 42 | CATSPER2P1 | cation channel sperm associated 2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:31054] | 10951 | 0.000 | -0.0498 | No |
| 43 | GOLM2 | golgi membrane protein 2 [Source:HGNC Symbol;Acc:HGNC:24892] | 10981 | 0.000 | -0.0506 | No |
| 44 | PLA2G4E-AS1 | PLA2G4E antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:51419] | 11537 | 0.000 | -0.0656 | No |
| 45 | PLA2G4F | phospholipase A2 group IVF [Source:HGNC Symbol;Acc:HGNC:27396] | 11744 | 0.000 | -0.0712 | No |
| 46 | PLA2G4D | phospholipase A2 group IVD [Source:HGNC Symbol;Acc:HGNC:30038] | 11745 | 0.000 | -0.0712 | No |
| 47 | PLA2G4E | phospholipase A2 group IVE [Source:HGNC Symbol;Acc:HGNC:24791] | 11746 | 0.000 | -0.0712 | No |
| 48 | PLA2G4B | phospholipase A2 group IVB [Source:HGNC Symbol;Acc:HGNC:9036] | 11747 | 0.000 | -0.0712 | No |
| 49 | CKMT1B | creatine kinase, mitochondrial 1B [Source:HGNC Symbol;Acc:HGNC:1995] | 12167 | 0.000 | -0.0825 | No |
| 50 | CKMT1A | creatine kinase, mitochondrial 1A [Source:HGNC Symbol;Acc:HGNC:31736] | 12237 | 0.000 | -0.0844 | No |
| 51 | MIR1282 | microRNA 1282 [Source:HGNC Symbol;Acc:HGNC:35360] | 12483 | 0.000 | -0.0910 | No |
| 52 | TGM7 | transglutaminase 7 [Source:HGNC Symbol;Acc:HGNC:30790] | 12528 | 0.000 | -0.0922 | No |
| 53 | TGM5 | transglutaminase 5 [Source:HGNC Symbol;Acc:HGNC:11781] | 12531 | 0.000 | -0.0922 | No |
| 54 | PPP1R14D | protein phosphatase 1 regulatory inhibitor subunit 14D [Source:HGNC Symbol;Acc:HGNC:14953] | 13095 | 0.000 | -0.1075 | No |
| 55 | SPTBN5 | spectrin beta, non-erythrocytic 5 [Source:HGNC Symbol;Acc:HGNC:15680] | 14257 | 0.000 | -0.1389 | No |
| 56 | PAK6 | p21 (RAC1) activated kinase 6 [Source:HGNC Symbol;Acc:HGNC:16061] | 14787 | 0.000 | -0.1532 | No |
| 57 | SPINT1 | serine peptidase inhibitor, Kunitz type 1 [Source:HGNC Symbol;Acc:HGNC:11246] | 15313 | 0.000 | -0.1674 | No |
| 58 | STRC | stereocilin [Source:HGNC Symbol;Acc:HGNC:16035] | 15520 | 0.000 | -0.1730 | No |
| 59 | KRT8P50 | keratin 8 pseudogene 50 [Source:HGNC Symbol;Acc:HGNC:48346] | 16501 | 0.000 | -0.1995 | No |
| 60 | PLCB2-AS1 | PLCB2 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:43662] | 16751 | 0.000 | -0.2062 | No |
| 61 | RNA5SP392 | RNA, 5S ribosomal pseudogene 392 [Source:HGNC Symbol;Acc:HGNC:43292] | 17069 | 0.000 | -0.2148 | No |
| 62 | RNA5SP393 | RNA, 5S ribosomal pseudogene 393 [Source:HGNC Symbol;Acc:HGNC:43293] | 17112 | 0.000 | -0.2160 | No |
| 63 | RNU6-610P | RNA, U6 small nuclear 610, pseudogene [Source:HGNC Symbol;Acc:HGNC:47573] | 17465 | 0.000 | -0.2255 | No |
| 64 | PDIA3P2 | protein disulfide isomerase family A member 3 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:49403] | 17489 | 0.000 | -0.2261 | No |
| 65 | PHGR1 | proline, histidine and glycine rich 1 [Source:HGNC Symbol;Acc:HGNC:37226] | 18375 | 0.000 | -0.2501 | No |
| 66 | HNRNPMP1 | heterogeneous nuclear ribonucleoprotein M pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:48745] | 18484 | 0.000 | -0.2530 | No |
| 67 | LTK | leukocyte receptor tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:6721] | 18814 | 0.000 | -0.2619 | No |
| 68 | RAD51-AS1 | RAD51 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:48621] | 19964 | 0.000 | -0.2930 | No |
| 69 | RNU6-354P | RNA, U6 small nuclear 354, pseudogene [Source:HGNC Symbol;Acc:HGNC:47317] | 20321 | 0.000 | -0.3026 | No |
| 70 | RNU6-353P | RNA, U6 small nuclear 353, pseudogene [Source:HGNC Symbol;Acc:HGNC:47316] | 20370 | 0.000 | -0.3039 | No |
| 71 | SRP14-AS1 | SRP14 antisense RNA1 (head to head) [Source:HGNC Symbol;Acc:HGNC:48619] | 20418 | 0.000 | -0.3052 | No |
| 72 | DISP2 | dispatched RND transporter family member 2 [Source:HGNC Symbol;Acc:HGNC:19712] | 20958 | 0.000 | -0.3198 | No |
| 73 | STRCP1 | stereocilin pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:33915] | 21145 | 0.000 | -0.3248 | No |
| 74 | EPB42 | erythrocyte membrane protein band 4.2 [Source:HGNC Symbol;Acc:HGNC:3381] | 21435 | 0.000 | -0.3326 | No |
| 75 | CCDC9B | coiled-coil domain containing 9B [Source:HGNC Symbol;Acc:HGNC:33488] | 21445 | 0.000 | -0.3329 | No |
| 76 | RN7SL487P | RNA, 7SL, cytoplasmic 487, pseudogene [Source:HGNC Symbol;Acc:HGNC:46503] | 21654 | 0.000 | -0.3385 | No |
| 77 | EIF4EBP2P2 | eukaryotic translation initiation factor 4E binding protein 2 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:49317] | 21933 | 0.000 | -0.3460 | No |
| 78 | RN7SL497P | RNA, 7SL, cytoplasmic 497, pseudogene [Source:HGNC Symbol;Acc:HGNC:46513] | 22107 | 0.000 | -0.3507 | No |
| 79 | MIR4310 | microRNA 4310 [Source:HGNC Symbol;Acc:HGNC:38383] | 22522 | 0.000 | -0.3619 | No |
| 80 | SPINT1-AS1 | SPINT1 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:53162] | 22780 | 0.000 | -0.3688 | No |
| 81 | RN7SL376P | RNA, 7SL, cytoplasmic 376, pseudogene [Source:HGNC Symbol;Acc:HGNC:46392] | 25301 | 0.000 | -0.4370 | No |
| 82 | MIR627 | microRNA 627 [Source:HGNC Symbol;Acc:HGNC:32883] | 27495 | 0.000 | -0.4964 | No |
| 83 | MIR626 | microRNA 626 [Source:HGNC Symbol;Acc:HGNC:32882] | 27496 | 0.000 | -0.4964 | No |
| 84 | INO80-AS1 | INO80 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:53138] | 27962 | 0.000 | -0.5090 | No |
| 85 | JMJD7-PLA2G4B | JMJD7-PLA2G4B readthrough [Source:HGNC Symbol;Acc:HGNC:34449] | 28826 | -0.077 | -0.5316 | No |
| 86 | H3P38 | H3 histone pseudogene 38 [Source:HGNC Symbol;Acc:HGNC:31028] | 28984 | -0.119 | -0.5347 | No |
| 87 | CCNDBP1 | cyclin D1 binding protein 1 [Source:HGNC Symbol;Acc:HGNC:1587] | 29652 | -0.277 | -0.5501 | No |
| 88 | INAFM2 | InaF motif containing 2 [Source:HGNC Symbol;Acc:HGNC:35165] | 29695 | -0.288 | -0.5484 | No |
| 89 | SNAP23 | synaptosome associated protein 23 [Source:HGNC Symbol;Acc:HGNC:11131] | 29710 | -0.293 | -0.5460 | No |
| 90 | FDPSP4 | farnesyl diphosphate synthase pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:3635] | 29837 | -0.318 | -0.5464 | No |
| 91 | CCDC32 | coiled-coil domain containing 32 [Source:HGNC Symbol;Acc:HGNC:28295] | 29894 | -0.330 | -0.5447 | No |
| 92 | ADAL | adenosine deaminase like [Source:HGNC Symbol;Acc:HGNC:31853] | 30156 | -0.397 | -0.5480 | No |
| 93 | RPAP1 | RNA polymerase II associated protein 1 [Source:HGNC Symbol;Acc:HGNC:24567] | 30857 | -0.581 | -0.5613 | Yes |
| 94 | CTDSPL2 | CTD small phosphatase like 2 [Source:HGNC Symbol;Acc:HGNC:26936] | 30910 | -0.592 | -0.5571 | Yes |
| 95 | OIP5 | Opa interacting protein 5 [Source:HGNC Symbol;Acc:HGNC:20300] | 31183 | -0.669 | -0.5580 | Yes |
| 96 | HAUS2 | HAUS augmin like complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:25530] | 31215 | -0.677 | -0.5523 | Yes |
| 97 | TTBK2 | tau tubulin kinase 2 [Source:HGNC Symbol;Acc:HGNC:19141] | 31227 | -0.681 | -0.5461 | Yes |
| 98 | ANKRD63 | ankyrin repeat domain 63 [Source:HGNC Symbol;Acc:HGNC:40027] | 31332 | -0.712 | -0.5421 | Yes |
| 99 | PPIP5K1 | diphosphoinositol pentakisphosphate kinase 1 [Source:HGNC Symbol;Acc:HGNC:29023] | 31404 | -0.733 | -0.5370 | Yes |
| 100 | CHP1 | calcineurin like EF-hand protein 1 [Source:HGNC Symbol;Acc:HGNC:17433] | 31453 | -0.750 | -0.5311 | Yes |
| 101 | WDR76 | WD repeat domain 76 [Source:HGNC Symbol;Acc:HGNC:25773] | 31589 | -0.768 | -0.5274 | Yes |
| 102 | CAPN3 | calpain 3 [Source:HGNC Symbol;Acc:HGNC:1480] | 31934 | -0.891 | -0.5281 | Yes |
| 103 | DNAJC17 | DnaJ heat shock protein family (Hsp40) member C17 [Source:HGNC Symbol;Acc:HGNC:25556] | 32059 | -0.925 | -0.5226 | Yes |
| 104 | CHST14 | carbohydrate sulfotransferase 14 [Source:HGNC Symbol;Acc:HGNC:24464] | 32067 | -0.930 | -0.5139 | Yes |
| 105 | EHD4 | EH domain containing 4 [Source:HGNC Symbol;Acc:HGNC:3245] | 32359 | -1.030 | -0.5119 | Yes |
| 106 | ZNF106 | zinc finger protein 106 [Source:HGNC Symbol;Acc:HGNC:12886] | 32389 | -1.038 | -0.5027 | Yes |
| 107 | DLL4 | delta like canonical Notch ligand 4 [Source:HGNC Symbol;Acc:HGNC:2910] | 32635 | -1.129 | -0.4985 | Yes |
| 108 | KNL1 | kinetochore scaffold 1 [Source:HGNC Symbol;Acc:HGNC:24054] | 32938 | -1.234 | -0.4948 | Yes |
| 109 | PLCB2 | phospholipase C beta 2 [Source:HGNC Symbol;Acc:HGNC:9055] | 32995 | -1.256 | -0.4843 | Yes |
| 110 | TUBGCP4 | tubulin gamma complex associated protein 4 [Source:HGNC Symbol;Acc:HGNC:16691] | 33002 | -1.259 | -0.4724 | Yes |
| 111 | VPS39 | VPS39 subunit of HOPS complex [Source:HGNC Symbol;Acc:HGNC:20593] | 33274 | -1.365 | -0.4666 | Yes |
| 112 | ZFYVE19 | zinc finger FYVE-type containing 19 [Source:HGNC Symbol;Acc:HGNC:20758] | 33432 | -1.436 | -0.4571 | Yes |
| 113 | RAD51 | RAD51 recombinase [Source:HGNC Symbol;Acc:HGNC:9817] | 33447 | -1.444 | -0.4436 | Yes |
| 114 | GAPDHP55 | glyceraldehyde 3 phosphate dehydrogenase pseudogene 55 [Source:HGNC Symbol;Acc:HGNC:38561] | 33524 | -1.451 | -0.4318 | Yes |
| 115 | SUMO2P15 | SUMO2 pseudogene 15 [Source:HGNC Symbol;Acc:HGNC:49353] | 33552 | -1.451 | -0.4186 | Yes |
| 116 | ITPKA | inositol-trisphosphate 3-kinase A [Source:HGNC Symbol;Acc:HGNC:6178] | 34165 | -1.696 | -0.4189 | Yes |
| 117 | OIP5-AS1 | OIP5 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:43563] | 34206 | -1.716 | -0.4035 | Yes |
| 118 | BNIP3P5 | BCL2 interacting protein 3 pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:39658] | 34249 | -1.742 | -0.3879 | Yes |
| 119 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 34488 | -1.854 | -0.3766 | Yes |
| 120 | IVD | isovaleryl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:6186] | 34702 | -1.970 | -0.3634 | Yes |
| 121 | MAPKBP1 | mitogen-activated protein kinase binding protein 1 [Source:HGNC Symbol;Acc:HGNC:29536] | 34715 | -1.978 | -0.3448 | Yes |
| 122 | CDAN1 | codanin 1 [Source:HGNC Symbol;Acc:HGNC:1713] | 35030 | -2.193 | -0.3322 | Yes |
| 123 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 35078 | -2.229 | -0.3121 | Yes |
| 124 | BUB1B-PAK6 | BUB1B-PAK6 readthrough [Source:HGNC Symbol;Acc:HGNC:52276] | 35098 | -2.244 | -0.2911 | Yes |
| 125 | EIF2AK4 | eukaryotic translation initiation factor 2 alpha kinase 4 [Source:HGNC Symbol;Acc:HGNC:19687] | 35462 | -2.443 | -0.2775 | Yes |
| 126 | GANC | glucosidase alpha, neutral C [Source:HGNC Symbol;Acc:HGNC:4139] | 35623 | -2.585 | -0.2570 | Yes |
| 127 | STARD9 | StAR related lipid transfer domain containing 9 [Source:HGNC Symbol;Acc:HGNC:19162] | 36062 | -2.988 | -0.2402 | Yes |
| 128 | CIBAR1P1 | CIBAR1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:32278] | 36129 | -3.078 | -0.2125 | Yes |
| 129 | JMJD7 | jumonji domain containing 7 [Source:HGNC Symbol;Acc:HGNC:34397] | 36173 | -3.137 | -0.1835 | Yes |
| 130 | KNSTRN | kinetochore localized astrin (SPAG5) binding protein [Source:HGNC Symbol;Acc:HGNC:30767] | 36395 | -3.379 | -0.1571 | Yes |
| 131 | MAP1A | microtubule associated protein 1A [Source:HGNC Symbol;Acc:HGNC:6835] | 36498 | -3.488 | -0.1264 | Yes |
| 132 | GPR176 | G protein-coupled receptor 176 [Source:HGNC Symbol;Acc:HGNC:32370] | 36500 | -3.490 | -0.0930 | Yes |
| 133 | BMF | Bcl2 modifying factor [Source:HGNC Symbol;Acc:HGNC:24132] | 36562 | -3.591 | -0.0602 | Yes |
| 134 | GCHFR | GTP cyclohydrolase I feedback regulator [Source:HGNC Symbol;Acc:HGNC:4194] | 36624 | -3.679 | -0.0265 | Yes |
| 135 | TYRO3 | TYRO3 protein tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:12446] | 36840 | -4.075 | 0.0068 | Yes |