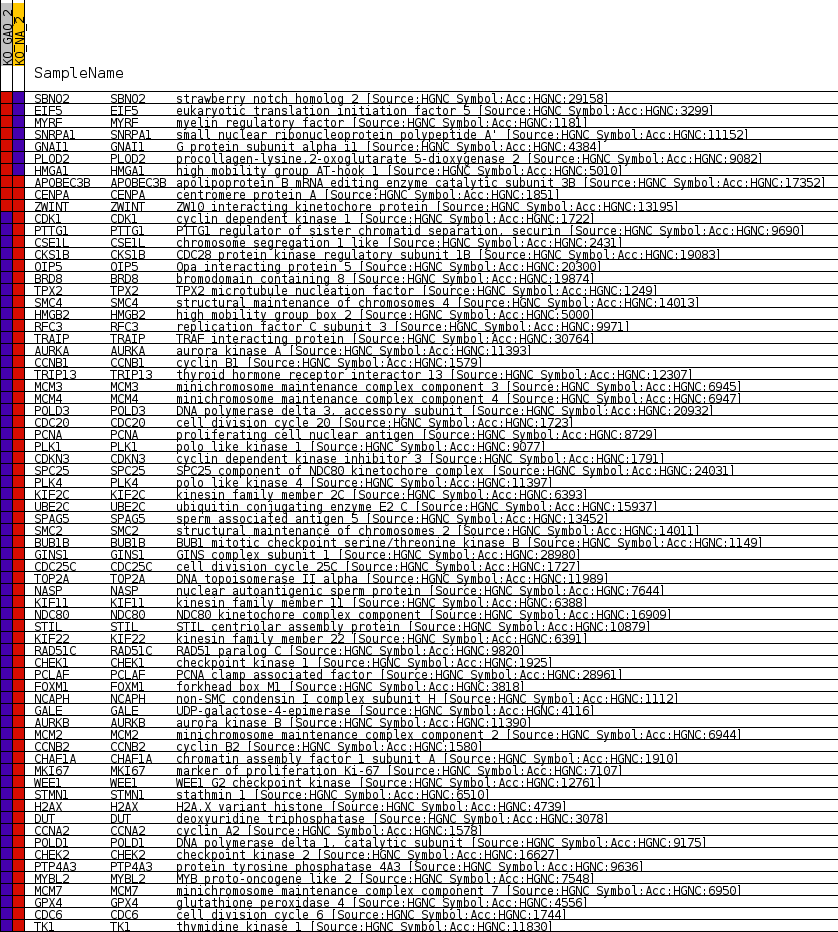
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
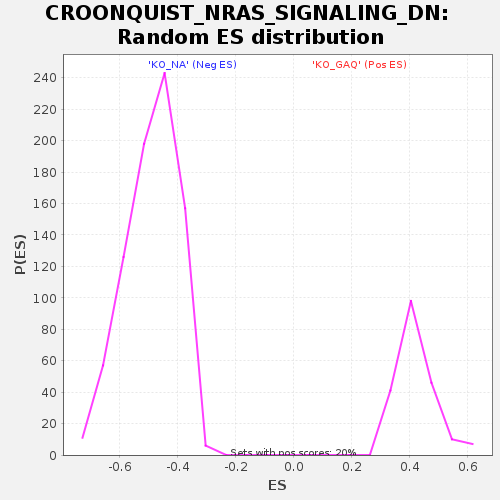
| GeneSet | CROONQUIST_NRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.80308276 |
| Normalized Enrichment Score (NES) | -1.6416236 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.013765815 |
| FWER p-Value | 0.249 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SBNO2 | strawberry notch homolog 2 [Source:HGNC Symbol;Acc:HGNC:29158] | 344 | 3.401 | 0.0172 | No |
| 2 | EIF5 | eukaryotic translation initiation factor 5 [Source:HGNC Symbol;Acc:HGNC:3299] | 2457 | 1.143 | -0.0310 | No |
| 3 | MYRF | myelin regulatory factor [Source:HGNC Symbol;Acc:HGNC:1181] | 3378 | 0.782 | -0.0497 | No |
| 4 | SNRPA1 | small nuclear ribonucleoprotein polypeptide A' [Source:HGNC Symbol;Acc:HGNC:11152] | 3609 | 0.700 | -0.0505 | No |
| 5 | GNAI1 | G protein subunit alpha i1 [Source:HGNC Symbol;Acc:HGNC:4384] | 4098 | 0.550 | -0.0594 | No |
| 6 | PLOD2 | procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 [Source:HGNC Symbol;Acc:HGNC:9082] | 4570 | 0.412 | -0.0689 | No |
| 7 | HMGA1 | high mobility group AT-hook 1 [Source:HGNC Symbol;Acc:HGNC:5010] | 4978 | 0.295 | -0.0776 | No |
| 8 | APOBEC3B | apolipoprotein B mRNA editing enzyme catalytic subunit 3B [Source:HGNC Symbol;Acc:HGNC:17352] | 12487 | 0.000 | -0.2804 | No |
| 9 | CENPA | centromere protein A [Source:HGNC Symbol;Acc:HGNC:1851] | 21259 | 0.000 | -0.5173 | No |
| 10 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 23551 | 0.000 | -0.5792 | No |
| 11 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 28955 | -0.111 | -0.7243 | No |
| 12 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 30002 | -0.356 | -0.7498 | No |
| 13 | CSE1L | chromosome segregation 1 like [Source:HGNC Symbol;Acc:HGNC:2431] | 30622 | -0.519 | -0.7625 | No |
| 14 | CKS1B | CDC28 protein kinase regulatory subunit 1B [Source:HGNC Symbol;Acc:HGNC:19083] | 31156 | -0.662 | -0.7717 | No |
| 15 | OIP5 | Opa interacting protein 5 [Source:HGNC Symbol;Acc:HGNC:20300] | 31183 | -0.669 | -0.7672 | No |
| 16 | BRD8 | bromodomain containing 8 [Source:HGNC Symbol;Acc:HGNC:19874] | 32512 | -1.089 | -0.7946 | Yes |
| 17 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 32541 | -1.098 | -0.7868 | Yes |
| 18 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 32582 | -1.110 | -0.7793 | Yes |
| 19 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 32901 | -1.221 | -0.7783 | Yes |
| 20 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 32933 | -1.233 | -0.7696 | Yes |
| 21 | TRAIP | TRAF interacting protein [Source:HGNC Symbol;Acc:HGNC:30764] | 32956 | -1.242 | -0.7605 | Yes |
| 22 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 33083 | -1.285 | -0.7539 | Yes |
| 23 | CCNB1 | cyclin B1 [Source:HGNC Symbol;Acc:HGNC:1579] | 33194 | -1.331 | -0.7465 | Yes |
| 24 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 33198 | -1.332 | -0.7362 | Yes |
| 25 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 33400 | -1.422 | -0.7306 | Yes |
| 26 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 33442 | -1.442 | -0.7205 | Yes |
| 27 | POLD3 | DNA polymerase delta 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:20932] | 33701 | -1.476 | -0.7159 | Yes |
| 28 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 33722 | -1.484 | -0.7049 | Yes |
| 29 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33797 | -1.517 | -0.6951 | Yes |
| 30 | PLK1 | polo like kinase 1 [Source:HGNC Symbol;Acc:HGNC:9077] | 33895 | -1.555 | -0.6856 | Yes |
| 31 | CDKN3 | cyclin dependent kinase inhibitor 3 [Source:HGNC Symbol;Acc:HGNC:1791] | 33991 | -1.612 | -0.6756 | Yes |
| 32 | SPC25 | SPC25 component of NDC80 kinetochore complex [Source:HGNC Symbol;Acc:HGNC:24031] | 34102 | -1.666 | -0.6657 | Yes |
| 33 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34155 | -1.691 | -0.6539 | Yes |
| 34 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 34210 | -1.718 | -0.6420 | Yes |
| 35 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 34312 | -1.770 | -0.6309 | Yes |
| 36 | SPAG5 | sperm associated antigen 5 [Source:HGNC Symbol;Acc:HGNC:13452] | 34343 | -1.784 | -0.6179 | Yes |
| 37 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 34487 | -1.852 | -0.6073 | Yes |
| 38 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 34488 | -1.854 | -0.5929 | Yes |
| 39 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 34533 | -1.882 | -0.5794 | Yes |
| 40 | CDC25C | cell division cycle 25C [Source:HGNC Symbol;Acc:HGNC:1727] | 34537 | -1.884 | -0.5648 | Yes |
| 41 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 34662 | -1.944 | -0.5530 | Yes |
| 42 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 34681 | -1.952 | -0.5383 | Yes |
| 43 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 34689 | -1.957 | -0.5233 | Yes |
| 44 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 34879 | -2.099 | -0.5121 | Yes |
| 45 | STIL | STIL centriolar assembly protein [Source:HGNC Symbol;Acc:HGNC:10879] | 34921 | -2.127 | -0.4966 | Yes |
| 46 | KIF22 | kinesin family member 22 [Source:HGNC Symbol;Acc:HGNC:6391] | 35069 | -2.220 | -0.4833 | Yes |
| 47 | RAD51C | RAD51 paralog C [Source:HGNC Symbol;Acc:HGNC:9820] | 35200 | -2.313 | -0.4688 | Yes |
| 48 | CHEK1 | checkpoint kinase 1 [Source:HGNC Symbol;Acc:HGNC:1925] | 35205 | -2.315 | -0.4509 | Yes |
| 49 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 35306 | -2.318 | -0.4355 | Yes |
| 50 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 35329 | -2.329 | -0.4180 | Yes |
| 51 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 35378 | -2.378 | -0.4008 | Yes |
| 52 | GALE | UDP-galactose-4-epimerase [Source:HGNC Symbol;Acc:HGNC:4116] | 35465 | -2.446 | -0.3841 | Yes |
| 53 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 35466 | -2.446 | -0.3650 | Yes |
| 54 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 35567 | -2.531 | -0.3480 | Yes |
| 55 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 35581 | -2.547 | -0.3286 | Yes |
| 56 | CHAF1A | chromatin assembly factor 1 subunit A [Source:HGNC Symbol;Acc:HGNC:1910] | 35614 | -2.578 | -0.3093 | Yes |
| 57 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 35643 | -2.607 | -0.2898 | Yes |
| 58 | WEE1 | WEE1 G2 checkpoint kinase [Source:HGNC Symbol;Acc:HGNC:12761] | 35705 | -2.646 | -0.2709 | Yes |
| 59 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 35813 | -2.748 | -0.2524 | Yes |
| 60 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 35814 | -2.749 | -0.2310 | Yes |
| 61 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 35839 | -2.770 | -0.2100 | Yes |
| 62 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 35850 | -2.788 | -0.1886 | Yes |
| 63 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36079 | -3.005 | -0.1714 | Yes |
| 64 | CHEK2 | checkpoint kinase 2 [Source:HGNC Symbol;Acc:HGNC:16627] | 36144 | -3.094 | -0.1490 | Yes |
| 65 | PTP4A3 | protein tyrosine phosphatase 4A3 [Source:HGNC Symbol;Acc:HGNC:9636] | 36323 | -3.270 | -0.1284 | Yes |
| 66 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36366 | -3.344 | -0.1035 | Yes |
| 67 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36496 | -3.483 | -0.0799 | Yes |
| 68 | GPX4 | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 36720 | -3.854 | -0.0559 | Yes |
| 69 | CDC6 | cell division cycle 6 [Source:HGNC Symbol;Acc:HGNC:1744] | 36809 | -4.009 | -0.0270 | Yes |
| 70 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 36977 | -4.446 | 0.0031 | Yes |