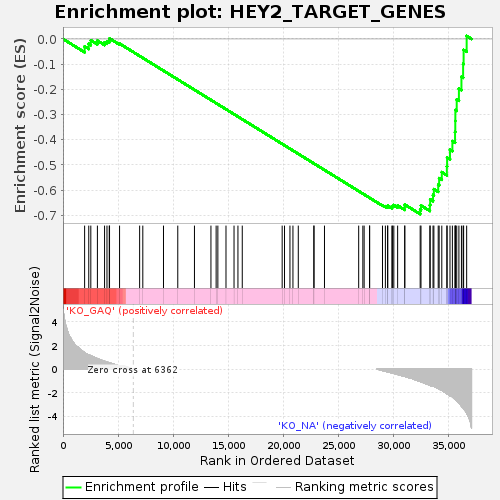
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HEY2_TARGET_GENES |
| Enrichment Score (ES) | -0.6953326 |
| Normalized Enrichment Score (NES) | -1.4140083 |
| Nominal p-value | 0.015247776 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | LINC01460 | long intergenic non-protein coding RNA 1460 [Source:HGNC Symbol;Acc:HGNC:50858] | 1963 | 1.378 | -0.0297 | No |
| 2 | ROCK1P1 | Rho associated coiled-coil containing protein kinase 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:37832] | 2338 | 1.205 | -0.0194 | No |
| 3 | EXOSC3 | exosome component 3 [Source:HGNC Symbol;Acc:HGNC:17944] | 2525 | 1.117 | -0.0055 | No |
| 4 | PRRC2A | proline rich coiled-coil 2A [Source:HGNC Symbol;Acc:HGNC:13918] | 3123 | 0.872 | -0.0068 | No |
| 5 | RBPJ | recombination signal binding protein for immunoglobulin kappa J region [Source:HGNC Symbol;Acc:HGNC:5724] | 3773 | 0.653 | -0.0133 | No |
| 6 | KLHDC4 | kelch domain containing 4 [Source:HGNC Symbol;Acc:HGNC:25272] | 3992 | 0.581 | -0.0093 | No |
| 7 | TMEM204 | transmembrane protein 204 [Source:HGNC Symbol;Acc:HGNC:14158] | 4198 | 0.518 | -0.0061 | No |
| 8 | GAS2L3 | growth arrest specific 2 like 3 [Source:HGNC Symbol;Acc:HGNC:27475] | 4224 | 0.512 | 0.0019 | No |
| 9 | NAA38 | N-alpha-acetyltransferase 38, NatC auxiliary subunit [Source:HGNC Symbol;Acc:HGNC:28212] | 5135 | 0.247 | -0.0184 | No |
| 10 | SDK1 | sidekick cell adhesion molecule 1 [Source:HGNC Symbol;Acc:HGNC:19307] | 6957 | 0.000 | -0.0676 | No |
| 11 | MYRIP | myosin VIIA and Rab interacting protein [Source:HGNC Symbol;Acc:HGNC:19156] | 7245 | 0.000 | -0.0754 | No |
| 12 | LINC01088 | long intergenic non-protein coding RNA 1088 [Source:HGNC Symbol;Acc:HGNC:49148] | 9122 | 0.000 | -0.1260 | No |
| 13 | CCDC162P | coiled-coil domain containing 162, pseudogene [Source:HGNC Symbol;Acc:HGNC:21565] | 10419 | 0.000 | -0.1611 | No |
| 14 | JAKMIP1 | janus kinase and microtubule interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:26460] | 11933 | 0.000 | -0.2019 | No |
| 15 | PDE1C | phosphodiesterase 1C [Source:HGNC Symbol;Acc:HGNC:8776] | 13430 | 0.000 | -0.2423 | No |
| 16 | LINC02248 | long intergenic non-protein coding RNA 2248 [Source:HGNC Symbol;Acc:HGNC:53147] | 13901 | 0.000 | -0.2550 | No |
| 17 | RUNX1T1 | RUNX1 partner transcriptional co-repressor 1 [Source:HGNC Symbol;Acc:HGNC:1535] | 14058 | 0.000 | -0.2592 | No |
| 18 | LINC02495 | long intergenic non-protein coding RNA 2495 [Source:HGNC Symbol;Acc:HGNC:27728] | 14796 | 0.000 | -0.2791 | No |
| 19 | MYBPC1 | myosin binding protein C1 [Source:HGNC Symbol;Acc:HGNC:7549] | 15526 | 0.000 | -0.2988 | No |
| 20 | IL23R | interleukin 23 receptor [Source:HGNC Symbol;Acc:HGNC:19100] | 15881 | 0.000 | -0.3084 | No |
| 21 | HDHD5-AS1 | HDHD5 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:1842] | 16273 | 0.000 | -0.3189 | No |
| 22 | RNA5S12 | RNA, 5S ribosomal 12 [Source:HGNC Symbol;Acc:HGNC:34373] | 19896 | 0.000 | -0.4168 | No |
| 23 | CELSR1 | cadherin EGF LAG seven-pass G-type receptor 1 [Source:HGNC Symbol;Acc:HGNC:1850] | 20109 | 0.000 | -0.4225 | No |
| 24 | MYLK3 | myosin light chain kinase 3 [Source:HGNC Symbol;Acc:HGNC:29826] | 20602 | 0.000 | -0.4358 | No |
| 25 | RBM24 | RNA binding motif protein 24 [Source:HGNC Symbol;Acc:HGNC:21539] | 20874 | 0.000 | -0.4431 | No |
| 26 | PDYN-AS1 | PDYN antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:53462] | 21359 | 0.000 | -0.4562 | No |
| 27 | PYGO1 | pygopus family PHD finger 1 [Source:HGNC Symbol;Acc:HGNC:30256] | 22773 | 0.000 | -0.4943 | No |
| 28 | RTN4R | reticulon 4 receptor [Source:HGNC Symbol;Acc:HGNC:18601] | 22785 | 0.000 | -0.4946 | No |
| 29 | LRRC14B | leucine rich repeat containing 14B [Source:HGNC Symbol;Acc:HGNC:37268] | 23737 | 0.000 | -0.5203 | No |
| 30 | ZNF563 | zinc finger protein 563 [Source:HGNC Symbol;Acc:HGNC:30498] | 26842 | 0.000 | -0.6042 | No |
| 31 | NUDT15 | nudix hydrolase 15 [Source:HGNC Symbol;Acc:HGNC:23063] | 27220 | 0.000 | -0.6143 | No |
| 32 | YAP1 | Yes1 associated transcriptional regulator [Source:HGNC Symbol;Acc:HGNC:16262] | 27337 | 0.000 | -0.6175 | No |
| 33 | MIR592 | microRNA 592 [Source:HGNC Symbol;Acc:HGNC:32848] | 27825 | 0.000 | -0.6306 | No |
| 34 | TCF7L1 | transcription factor 7 like 1 [Source:HGNC Symbol;Acc:HGNC:11640] | 27841 | 0.000 | -0.6310 | No |
| 35 | CCNC | cyclin C [Source:HGNC Symbol;Acc:HGNC:1581] | 29003 | -0.123 | -0.6603 | No |
| 36 | AMPD3 | adenosine monophosphate deaminase 3 [Source:HGNC Symbol;Acc:HGNC:470] | 29265 | -0.190 | -0.6641 | No |
| 37 | RPS7 | ribosomal protein S7 [Source:HGNC Symbol;Acc:HGNC:10440] | 29466 | -0.230 | -0.6656 | No |
| 38 | PRUNE2 | prune homolog 2 with BCH domain [Source:HGNC Symbol;Acc:HGNC:25209] | 29481 | -0.235 | -0.6620 | No |
| 39 | TMEM70 | transmembrane protein 70 [Source:HGNC Symbol;Acc:HGNC:26050] | 29855 | -0.320 | -0.6667 | No |
| 40 | HERC1 | HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 [Source:HGNC Symbol;Acc:HGNC:4867] | 29920 | -0.336 | -0.6627 | No |
| 41 | AIMP2 | aminoacyl tRNA synthetase complex interacting multifunctional protein 2 [Source:HGNC Symbol;Acc:HGNC:20609] | 30032 | -0.361 | -0.6596 | No |
| 42 | PTMA | prothymosin alpha [Source:HGNC Symbol;Acc:HGNC:9623] | 30378 | -0.455 | -0.6612 | No |
| 43 | FIP1L1 | factor interacting with PAPOLA and CPSF1 [Source:HGNC Symbol;Acc:HGNC:19124] | 31024 | -0.620 | -0.6681 | No |
| 44 | RPL24 | ribosomal protein L24 [Source:HGNC Symbol;Acc:HGNC:10325] | 31025 | -0.621 | -0.6576 | No |
| 45 | TMCC2 | transmembrane and coiled-coil domain family 2 [Source:HGNC Symbol;Acc:HGNC:24239] | 32424 | -1.053 | -0.6775 | Yes |
| 46 | E2F6 | E2F transcription factor 6 [Source:HGNC Symbol;Acc:HGNC:3120] | 32498 | -1.083 | -0.6611 | Yes |
| 47 | MTCH1 | mitochondrial carrier 1 [Source:HGNC Symbol;Acc:HGNC:17586] | 33303 | -1.376 | -0.6595 | Yes |
| 48 | PPP1R12B | protein phosphatase 1 regulatory subunit 12B [Source:HGNC Symbol;Acc:HGNC:7619] | 33333 | -1.387 | -0.6368 | Yes |
| 49 | CDS1 | CDP-diacylglycerol synthase 1 [Source:HGNC Symbol;Acc:HGNC:1800] | 33591 | -1.451 | -0.6191 | Yes |
| 50 | ARNT2 | aryl hydrocarbon receptor nuclear translocator 2 [Source:HGNC Symbol;Acc:HGNC:16876] | 33668 | -1.464 | -0.5964 | Yes |
| 51 | STIM1 | stromal interaction molecule 1 [Source:HGNC Symbol;Acc:HGNC:11386] | 34061 | -1.647 | -0.5791 | Yes |
| 52 | CDK5R1 | cyclin dependent kinase 5 regulatory subunit 1 [Source:HGNC Symbol;Acc:HGNC:1775] | 34146 | -1.687 | -0.5527 | Yes |
| 53 | HDHD5 | haloacid dehalogenase like hydrolase domain containing 5 [Source:HGNC Symbol;Acc:HGNC:1843] | 34387 | -1.799 | -0.5287 | Yes |
| 54 | SMIM8 | small integral membrane protein 8 [Source:HGNC Symbol;Acc:HGNC:21401] | 34846 | -2.079 | -0.5059 | Yes |
| 55 | PMS2 | PMS1 homolog 2, mismatch repair system component [Source:HGNC Symbol;Acc:HGNC:9122] | 34869 | -2.092 | -0.4710 | Yes |
| 56 | PIK3IP1 | phosphoinositide-3-kinase interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:24942] | 35130 | -2.271 | -0.4396 | Yes |
| 57 | ACSL1 | acyl-CoA synthetase long chain family member 1 [Source:HGNC Symbol;Acc:HGNC:3569] | 35348 | -2.344 | -0.4057 | Yes |
| 58 | COQ8A | coenzyme Q8A [Source:HGNC Symbol;Acc:HGNC:16812] | 35591 | -2.555 | -0.3690 | Yes |
| 59 | EML6 | EMAP like 6 [Source:HGNC Symbol;Acc:HGNC:35412] | 35612 | -2.573 | -0.3259 | Yes |
| 60 | MTA3 | metastasis associated 1 family member 3 [Source:HGNC Symbol;Acc:HGNC:23784] | 35618 | -2.579 | -0.2823 | Yes |
| 61 | TNK2 | tyrosine kinase non receptor 2 [Source:HGNC Symbol;Acc:HGNC:19297] | 35741 | -2.677 | -0.2402 | Yes |
| 62 | TANC1 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 [Source:HGNC Symbol;Acc:HGNC:29364] | 35940 | -2.854 | -0.1972 | Yes |
| 63 | IFT140 | intraflagellar transport 140 [Source:HGNC Symbol;Acc:HGNC:29077] | 36170 | -3.134 | -0.1503 | Yes |
| 64 | PTP4A3 | protein tyrosine phosphatase 4A3 [Source:HGNC Symbol;Acc:HGNC:9636] | 36323 | -3.270 | -0.0990 | Yes |
| 65 | FANCA | FA complementation group A [Source:HGNC Symbol;Acc:HGNC:3582] | 36368 | -3.349 | -0.0434 | Yes |
| 66 | LPIN1 | lipin 1 [Source:HGNC Symbol;Acc:HGNC:13345] | 36644 | -3.711 | 0.0120 | Yes |