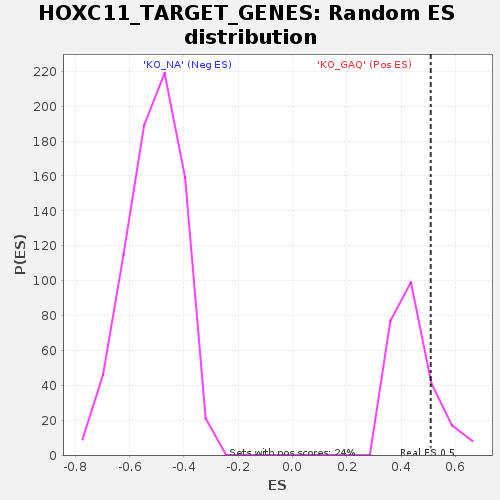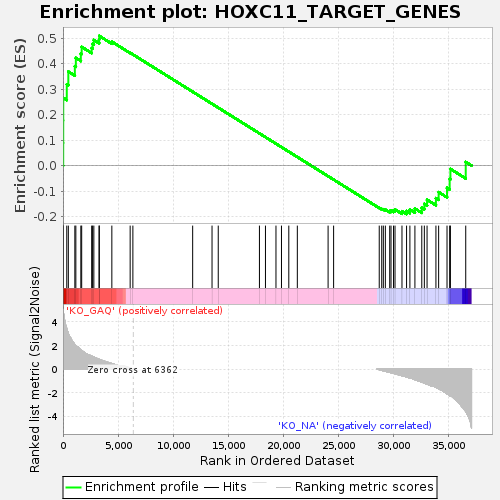
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_GAQ |
| GeneSet | HOXC11_TARGET_GENES |
| Enrichment Score (ES) | 0.5094138 |
| Normalized Enrichment Score (NES) | 1.1511655 |
| Nominal p-value | 0.16528925 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PTGS2 | prostaglandin-endoperoxide synthase 2 [Source:HGNC Symbol;Acc:HGNC:9605] | 18 | 4.924 | 0.0886 | Yes |
| 2 | CCL5 | C-C motif chemokine ligand 5 [Source:HGNC Symbol;Acc:HGNC:10632] | 20 | 4.911 | 0.1774 | Yes |
| 3 | IL10 | interleukin 10 [Source:HGNC Symbol;Acc:HGNC:5962] | 22 | 4.908 | 0.2662 | Yes |
| 4 | ERCC1 | ERCC excision repair 1, endonuclease non-catalytic subunit [Source:HGNC Symbol;Acc:HGNC:3433] | 343 | 3.403 | 0.3191 | Yes |
| 5 | PDAP1 | PDGFA associated protein 1 [Source:HGNC Symbol;Acc:HGNC:14634] | 484 | 2.975 | 0.3691 | Yes |
| 6 | TRMT13 | tRNA methyltransferase 13 homolog [Source:HGNC Symbol;Acc:HGNC:25502] | 1075 | 2.055 | 0.3904 | Yes |
| 7 | SASS6 | SAS-6 centriolar assembly protein [Source:HGNC Symbol;Acc:HGNC:25403] | 1166 | 1.938 | 0.4230 | Yes |
| 8 | SLC3A2 | solute carrier family 3 member 2 [Source:HGNC Symbol;Acc:HGNC:11026] | 1619 | 1.616 | 0.4400 | Yes |
| 9 | CPEB4 | cytoplasmic polyadenylation element binding protein 4 [Source:HGNC Symbol;Acc:HGNC:21747] | 1696 | 1.562 | 0.4663 | Yes |
| 10 | NIF3L1 | NGG1 interacting factor 3 like 1 [Source:HGNC Symbol;Acc:HGNC:13390] | 2603 | 1.084 | 0.4614 | Yes |
| 11 | CACYBP | calcyclin binding protein [Source:HGNC Symbol;Acc:HGNC:30423] | 2686 | 1.053 | 0.4783 | Yes |
| 12 | USO1 | USO1 vesicle transport factor [Source:HGNC Symbol;Acc:HGNC:30904] | 2800 | 1.007 | 0.4934 | Yes |
| 13 | GARS1 | glycyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:4162] | 3278 | 0.811 | 0.4952 | Yes |
| 14 | ESR1 | estrogen receptor 1 [Source:HGNC Symbol;Acc:HGNC:3467] | 3291 | 0.804 | 0.5094 | Yes |
| 15 | RAB5B | RAB5B, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9784] | 4434 | 0.449 | 0.4867 | No |
| 16 | ISY1 | ISY1 splicing factor homolog [Source:HGNC Symbol;Acc:HGNC:29201] | 6096 | 0.070 | 0.4431 | No |
| 17 | PPIL3 | peptidylprolyl isomerase like 3 [Source:HGNC Symbol;Acc:HGNC:9262] | 6343 | 0.003 | 0.4365 | No |
| 18 | S100B | S100 calcium binding protein B [Source:HGNC Symbol;Acc:HGNC:10500] | 11771 | 0.000 | 0.2900 | No |
| 19 | ELAVL4 | ELAV like RNA binding protein 4 [Source:HGNC Symbol;Acc:HGNC:3315] | 13529 | 0.000 | 0.2426 | No |
| 20 | MTCO3P12 | MT-CO3 pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:52042] | 14093 | 0.000 | 0.2274 | No |
| 21 | ISY1-RAB43 | ISY1-RAB43 readthrough [Source:HGNC Symbol;Acc:HGNC:42969] | 17832 | 0.000 | 0.1265 | No |
| 22 | CCNB1IP1 | cyclin B1 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:19437] | 18379 | 0.000 | 0.1118 | No |
| 23 | GCNT3 | glucosaminyl (N-acetyl) transferase 3, mucin type [Source:HGNC Symbol;Acc:HGNC:4205] | 19333 | 0.000 | 0.0860 | No |
| 24 | TBX3 | T-box transcription factor 3 [Source:HGNC Symbol;Acc:HGNC:11602] | 19833 | 0.000 | 0.0726 | No |
| 25 | PACERR | PTGS2 antisense NFKB1 complex-mediated expression regulator RNA [Source:HGNC Symbol;Acc:HGNC:50552] | 20498 | 0.000 | 0.0546 | No |
| 26 | MTND6P4 | MT-ND6 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:39467] | 21272 | 0.000 | 0.0338 | No |
| 27 | MIR34AHG | MIR34A host gene [Source:HGNC Symbol;Acc:HGNC:51913] | 24065 | 0.000 | -0.0416 | No |
| 28 | TIGD4 | tigger transposable element derived 4 [Source:HGNC Symbol;Acc:HGNC:18335] | 24566 | 0.000 | -0.0551 | No |
| 29 | DNAJB4 | DnaJ heat shock protein family (Hsp40) member B4 [Source:HGNC Symbol;Acc:HGNC:14886] | 28708 | -0.050 | -0.1660 | No |
| 30 | EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 [Source:HGNC Symbol;Acc:HGNC:3189] | 28934 | -0.106 | -0.1701 | No |
| 31 | MRPL33 | mitochondrial ribosomal protein L33 [Source:HGNC Symbol;Acc:HGNC:14487] | 29079 | -0.145 | -0.1714 | No |
| 32 | TCP1 | t-complex 1 [Source:HGNC Symbol;Acc:HGNC:11655] | 29262 | -0.190 | -0.1729 | No |
| 33 | TPR | translocated promoter region, nuclear basket protein [Source:HGNC Symbol;Acc:HGNC:12017] | 29659 | -0.280 | -0.1785 | No |
| 34 | RPL7 | ribosomal protein L7 [Source:HGNC Symbol;Acc:HGNC:10363] | 29777 | -0.308 | -0.1761 | No |
| 35 | GPSM3 | G protein signaling modulator 3 [Source:HGNC Symbol;Acc:HGNC:13945] | 30005 | -0.356 | -0.1758 | No |
| 36 | MTCYBP35 | MT-CYB pseudogene 35 [Source:HGNC Symbol;Acc:HGNC:52303] | 30140 | -0.392 | -0.1723 | No |
| 37 | CEP85 | centrosomal protein 85 [Source:HGNC Symbol;Acc:HGNC:25309] | 30772 | -0.559 | -0.1792 | No |
| 38 | ACBD6 | acyl-CoA binding domain containing 6 [Source:HGNC Symbol;Acc:HGNC:23339] | 31189 | -0.670 | -0.1783 | No |
| 39 | CENPL | centromere protein L [Source:HGNC Symbol;Acc:HGNC:17879] | 31491 | -0.761 | -0.1727 | No |
| 40 | INTS4 | integrator complex subunit 4 [Source:HGNC Symbol;Acc:HGNC:25048] | 31946 | -0.894 | -0.1688 | No |
| 41 | PMEL | premelanosome protein [Source:HGNC Symbol;Acc:HGNC:10880] | 32570 | -1.107 | -0.1656 | No |
| 42 | CCDC14 | coiled-coil domain containing 14 [Source:HGNC Symbol;Acc:HGNC:25766] | 32802 | -1.183 | -0.1504 | No |
| 43 | MRPL18 | mitochondrial ribosomal protein L18 [Source:HGNC Symbol;Acc:HGNC:14477] | 33049 | -1.275 | -0.1340 | No |
| 44 | GGCX | gamma-glutamyl carboxylase [Source:HGNC Symbol;Acc:HGNC:4247] | 33854 | -1.540 | -0.1278 | No |
| 45 | DUSP6 | dual specificity phosphatase 6 [Source:HGNC Symbol;Acc:HGNC:3072] | 34100 | -1.666 | -0.1043 | No |
| 46 | GCDH | glutaryl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4189] | 34861 | -2.086 | -0.0871 | No |
| 47 | DARS2 | aspartyl-tRNA synthetase 2, mitochondrial [Source:HGNC Symbol;Acc:HGNC:25538] | 35101 | -2.245 | -0.0529 | No |
| 48 | TERT | telomerase reverse transcriptase [Source:HGNC Symbol;Acc:HGNC:11730] | 35169 | -2.301 | -0.0131 | No |
| 49 | NAV1 | neuron navigator 1 [Source:HGNC Symbol;Acc:HGNC:15989] | 36560 | -3.589 | 0.0143 | No |