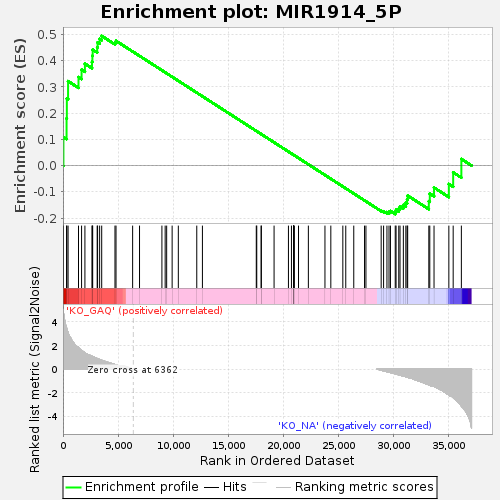
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_GAQ |
| GeneSet | MIR1914_5P |
| Enrichment Score (ES) | 0.4941915 |
| Normalized Enrichment Score (NES) | 1.1798334 |
| Nominal p-value | 0.08450704 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FLRT3 | fibronectin leucine rich transmembrane protein 3 [Source:HGNC Symbol;Acc:HGNC:3762] | 39 | 4.814 | 0.1086 | Yes |
| 2 | ZNF730 | zinc finger protein 730 [Source:HGNC Symbol;Acc:HGNC:32470] | 324 | 3.462 | 0.1798 | Yes |
| 3 | RAPGEF5 | Rap guanine nucleotide exchange factor 5 [Source:HGNC Symbol;Acc:HGNC:16862] | 348 | 3.365 | 0.2558 | Yes |
| 4 | TOR1AIP2 | torsin 1A interacting protein 2 [Source:HGNC Symbol;Acc:HGNC:24055] | 461 | 3.032 | 0.3218 | Yes |
| 5 | ST18 | ST18 C2H2C-type zinc finger transcription factor [Source:HGNC Symbol;Acc:HGNC:18695] | 1407 | 1.797 | 0.3372 | Yes |
| 6 | TRIM14 | tripartite motif containing 14 [Source:HGNC Symbol;Acc:HGNC:16283] | 1686 | 1.571 | 0.3655 | Yes |
| 7 | UBE2Z | ubiquitin conjugating enzyme E2 Z [Source:HGNC Symbol;Acc:HGNC:25847] | 1989 | 1.362 | 0.3883 | Yes |
| 8 | EXOC5 | exocyst complex component 5 [Source:HGNC Symbol;Acc:HGNC:10696] | 2633 | 1.072 | 0.3954 | Yes |
| 9 | KBTBD2 | kelch repeat and BTB domain containing 2 [Source:HGNC Symbol;Acc:HGNC:21751] | 2668 | 1.058 | 0.4186 | Yes |
| 10 | MGA | MAX dimerization protein MGA [Source:HGNC Symbol;Acc:HGNC:14010] | 2695 | 1.050 | 0.4418 | Yes |
| 11 | SPSB1 | splA/ryanodine receptor domain and SOCS box containing 1 [Source:HGNC Symbol;Acc:HGNC:30628] | 3103 | 0.881 | 0.4508 | Yes |
| 12 | DHRS3 | dehydrogenase/reductase 3 [Source:HGNC Symbol;Acc:HGNC:17693] | 3140 | 0.865 | 0.4696 | Yes |
| 13 | DNAJA2 | DnaJ heat shock protein family (Hsp40) member A2 [Source:HGNC Symbol;Acc:HGNC:14884] | 3325 | 0.791 | 0.4826 | Yes |
| 14 | SLC33A1 | solute carrier family 33 member 1 [Source:HGNC Symbol;Acc:HGNC:95] | 3513 | 0.730 | 0.4942 | Yes |
| 15 | RBMS1 | RNA binding motif single stranded interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:9907] | 4718 | 0.369 | 0.4701 | No |
| 16 | TOMM5 | translocase of outer mitochondrial membrane 5 [Source:HGNC Symbol;Acc:HGNC:31369] | 4813 | 0.342 | 0.4753 | No |
| 17 | BCAP29 | B cell receptor associated protein 29 [Source:HGNC Symbol;Acc:HGNC:24131] | 6321 | 0.009 | 0.4348 | No |
| 18 | ZNF98 | zinc finger protein 98 [Source:HGNC Symbol;Acc:HGNC:13174] | 6947 | 0.000 | 0.4179 | No |
| 19 | HIF3A | hypoxia inducible factor 3 subunit alpha [Source:HGNC Symbol;Acc:HGNC:15825] | 8979 | 0.000 | 0.3631 | No |
| 20 | DPYSL5 | dihydropyrimidinase like 5 [Source:HGNC Symbol;Acc:HGNC:20637] | 9274 | 0.000 | 0.3552 | No |
| 21 | ZNF138 | zinc finger protein 138 [Source:HGNC Symbol;Acc:HGNC:12922] | 9410 | 0.000 | 0.3515 | No |
| 22 | KCNQ4 | potassium voltage-gated channel subfamily Q member 4 [Source:HGNC Symbol;Acc:HGNC:6298] | 9907 | 0.000 | 0.3381 | No |
| 23 | EMX2 | empty spiracles homeobox 2 [Source:HGNC Symbol;Acc:HGNC:3341] | 10469 | 0.000 | 0.3230 | No |
| 24 | NCAM1 | neural cell adhesion molecule 1 [Source:HGNC Symbol;Acc:HGNC:7656] | 12143 | 0.000 | 0.2778 | No |
| 25 | CDKN2B | cyclin dependent kinase inhibitor 2B [Source:HGNC Symbol;Acc:HGNC:1788] | 12656 | 0.000 | 0.2639 | No |
| 26 | GPR39 | G protein-coupled receptor 39 [Source:HGNC Symbol;Acc:HGNC:4496] | 17545 | 0.000 | 0.1319 | No |
| 27 | SH2D7 | SH2 domain containing 7 [Source:HGNC Symbol;Acc:HGNC:34549] | 17589 | 0.000 | 0.1308 | No |
| 28 | SYDE1 | synapse defective Rho GTPase homolog 1 [Source:HGNC Symbol;Acc:HGNC:25824] | 17984 | 0.000 | 0.1201 | No |
| 29 | CASKIN1 | CASK interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:20879] | 18006 | 0.000 | 0.1196 | No |
| 30 | SHISAL1 | shisa like 1 [Source:HGNC Symbol;Acc:HGNC:29335] | 19161 | 0.000 | 0.0884 | No |
| 31 | ATP1B4 | ATPase Na+/K+ transporting family member beta 4 [Source:HGNC Symbol;Acc:HGNC:808] | 20465 | 0.000 | 0.0532 | No |
| 32 | NELL2 | neural EGFL like 2 [Source:HGNC Symbol;Acc:HGNC:7751] | 20738 | 0.000 | 0.0458 | No |
| 33 | TSPY4 | testis specific protein Y-linked 4 [Source:HGNC Symbol;Acc:HGNC:37287] | 20925 | 0.000 | 0.0408 | No |
| 34 | TSPY2 | testis specific protein Y-linked 2 [Source:HGNC Symbol;Acc:HGNC:23924] | 20926 | 0.000 | 0.0408 | No |
| 35 | TSPY3 | testis specific protein Y-linked 3 [Source:HGNC Symbol;Acc:HGNC:33876] | 20927 | 0.000 | 0.0408 | No |
| 36 | TSPY1 | testis specific protein Y-linked 1 [Source:HGNC Symbol;Acc:HGNC:12381] | 20928 | 0.000 | 0.0408 | No |
| 37 | TSPY8 | testis specific protein Y-linked 8 [Source:HGNC Symbol;Acc:HGNC:37471] | 20931 | 0.000 | 0.0408 | No |
| 38 | CNGA3 | cyclic nucleotide gated channel subunit alpha 3 [Source:HGNC Symbol;Acc:HGNC:2150] | 21005 | 0.000 | 0.0388 | No |
| 39 | MYO16 | myosin XVI [Source:HGNC Symbol;Acc:HGNC:29822] | 21380 | 0.000 | 0.0287 | No |
| 40 | PLAAT2 | phospholipase A and acyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:17824] | 22274 | 0.000 | 0.0046 | No |
| 41 | TSPY10 | testis specific protein Y-linked 10 [Source:HGNC Symbol;Acc:HGNC:37473] | 23780 | 0.000 | -0.0361 | No |
| 42 | ZBTB10 | zinc finger and BTB domain containing 10 [Source:HGNC Symbol;Acc:HGNC:30953] | 24313 | 0.000 | -0.0504 | No |
| 43 | DUSP26 | dual specificity phosphatase 26 [Source:HGNC Symbol;Acc:HGNC:28161] | 25403 | 0.000 | -0.0799 | No |
| 44 | ADAMTS15 | ADAM metallopeptidase with thrombospondin type 1 motif 15 [Source:HGNC Symbol;Acc:HGNC:16305] | 25669 | 0.000 | -0.0870 | No |
| 45 | ZNF675 | zinc finger protein 675 [Source:HGNC Symbol;Acc:HGNC:30768] | 26395 | 0.000 | -0.1066 | No |
| 46 | TBC1D30 | TBC1 domain family member 30 [Source:HGNC Symbol;Acc:HGNC:29164] | 27372 | 0.000 | -0.1329 | No |
| 47 | PHEX | phosphate regulating endopeptidase homolog X-linked [Source:HGNC Symbol;Acc:HGNC:8918] | 27500 | 0.000 | -0.1364 | No |
| 48 | ZNF273 | zinc finger protein 273 [Source:HGNC Symbol;Acc:HGNC:13067] | 28883 | -0.095 | -0.1716 | No |
| 49 | ACADSB | acyl-CoA dehydrogenase short/branched chain [Source:HGNC Symbol;Acc:HGNC:91] | 29103 | -0.147 | -0.1741 | No |
| 50 | CALD1 | caldesmon 1 [Source:HGNC Symbol;Acc:HGNC:1441] | 29407 | -0.218 | -0.1773 | No |
| 51 | ZNF680 | zinc finger protein 680 [Source:HGNC Symbol;Acc:HGNC:26897] | 29595 | -0.262 | -0.1764 | No |
| 52 | SKIDA1 | SKI/DACH domain containing 1 [Source:HGNC Symbol;Acc:HGNC:32697] | 29726 | -0.297 | -0.1732 | No |
| 53 | CLN8 | CLN8 transmembrane ER and ERGIC protein [Source:HGNC Symbol;Acc:HGNC:2079] | 30160 | -0.398 | -0.1758 | No |
| 54 | IBA57 | iron-sulfur cluster assembly factor IBA57 [Source:HGNC Symbol;Acc:HGNC:27302] | 30235 | -0.416 | -0.1683 | No |
| 55 | CCDC57 | coiled-coil domain containing 57 [Source:HGNC Symbol;Acc:HGNC:27564] | 30481 | -0.479 | -0.1640 | No |
| 56 | BMPR1A | bone morphogenetic protein receptor type 1A [Source:HGNC Symbol;Acc:HGNC:1076] | 30594 | -0.511 | -0.1554 | No |
| 57 | TCF20 | transcription factor 20 [Source:HGNC Symbol;Acc:HGNC:11631] | 30897 | -0.589 | -0.1502 | No |
| 58 | ATP2C1 | ATPase secretory pathway Ca2+ transporting 1 [Source:HGNC Symbol;Acc:HGNC:13211] | 31123 | -0.649 | -0.1415 | No |
| 59 | FOXP2 | forkhead box P2 [Source:HGNC Symbol;Acc:HGNC:13875] | 31238 | -0.685 | -0.1290 | No |
| 60 | SEPHS1 | selenophosphate synthetase 1 [Source:HGNC Symbol;Acc:HGNC:19685] | 31289 | -0.700 | -0.1144 | No |
| 61 | CEP350 | centrosomal protein 350 [Source:HGNC Symbol;Acc:HGNC:24238] | 33209 | -1.335 | -0.1358 | No |
| 62 | ZADH2 | zinc binding alcohol dehydrogenase domain containing 2 [Source:HGNC Symbol;Acc:HGNC:28697] | 33297 | -1.374 | -0.1069 | No |
| 63 | SATB2 | SATB homeobox 2 [Source:HGNC Symbol;Acc:HGNC:21637] | 33682 | -1.469 | -0.0838 | No |
| 64 | RAP2A | RAP2A, member of RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9861] | 35027 | -2.192 | -0.0702 | No |
| 65 | ZNF112 | zinc finger protein 112 [Source:HGNC Symbol;Acc:HGNC:12892] | 35420 | -2.406 | -0.0260 | No |
| 66 | KCNC4 | potassium voltage-gated channel subfamily C member 4 [Source:HGNC Symbol;Acc:HGNC:6236] | 36163 | -3.119 | 0.0250 | No |

