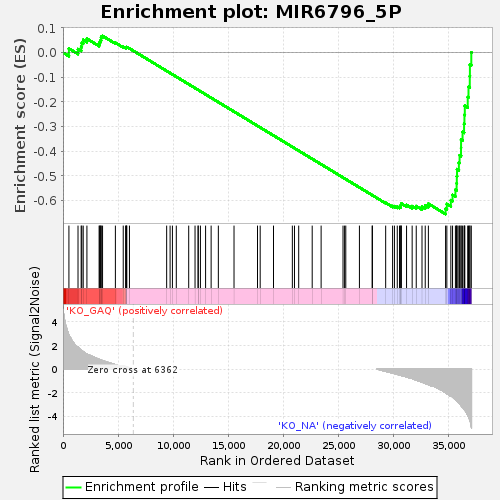
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | MIR6796_5P |
| Enrichment Score (ES) | -0.6544541 |
| Normalized Enrichment Score (NES) | -1.3669221 |
| Nominal p-value | 0.021197008 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | TXNRD1 | thioredoxin reductase 1 [Source:HGNC Symbol;Acc:HGNC:12437] | 537 | 2.855 | 0.0164 | No |
| 2 | MSN | moesin [Source:HGNC Symbol;Acc:HGNC:7373] | 1363 | 1.854 | 0.0142 | No |
| 3 | PDK4 | pyruvate dehydrogenase kinase 4 [Source:HGNC Symbol;Acc:HGNC:8812] | 1648 | 1.596 | 0.0238 | No |
| 4 | CRISPLD2 | cysteine rich secretory protein LCCL domain containing 2 [Source:HGNC Symbol;Acc:HGNC:25248] | 1706 | 1.553 | 0.0391 | No |
| 5 | MDK | midkine [Source:HGNC Symbol;Acc:HGNC:6972] | 1835 | 1.471 | 0.0516 | No |
| 6 | HBEGF | heparin binding EGF like growth factor [Source:HGNC Symbol;Acc:HGNC:3059] | 2171 | 1.253 | 0.0561 | No |
| 7 | GARS1 | glycyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:4162] | 3278 | 0.811 | 0.0350 | No |
| 8 | DNAJA2 | DnaJ heat shock protein family (Hsp40) member A2 [Source:HGNC Symbol;Acc:HGNC:14884] | 3325 | 0.791 | 0.0423 | No |
| 9 | BSDC1 | BSD domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25501] | 3393 | 0.777 | 0.0489 | No |
| 10 | AKT1S1 | AKT1 substrate 1 [Source:HGNC Symbol;Acc:HGNC:28426] | 3456 | 0.752 | 0.0554 | No |
| 11 | ELOVL1 | ELOVL fatty acid elongase 1 [Source:HGNC Symbol;Acc:HGNC:14418] | 3457 | 0.752 | 0.0636 | No |
| 12 | KMT2B | lysine methyltransferase 2B [Source:HGNC Symbol;Acc:HGNC:15840] | 3591 | 0.707 | 0.0676 | No |
| 13 | ARF3 | ADP ribosylation factor 3 [Source:HGNC Symbol;Acc:HGNC:654] | 4755 | 0.357 | 0.0401 | No |
| 14 | AUNIP | aurora kinase A and ninein interacting protein [Source:HGNC Symbol;Acc:HGNC:28363] | 5468 | 0.239 | 0.0234 | No |
| 15 | PARVA | parvin alpha [Source:HGNC Symbol;Acc:HGNC:14652] | 5686 | 0.193 | 0.0196 | No |
| 16 | SCYL3 | SCY1 like pseudokinase 3 [Source:HGNC Symbol;Acc:HGNC:19285] | 5731 | 0.181 | 0.0204 | No |
| 17 | SETD5 | SET domain containing 5 [Source:HGNC Symbol;Acc:HGNC:25566] | 5767 | 0.169 | 0.0213 | No |
| 18 | PPP1R2 | protein phosphatase 1 regulatory inhibitor subunit 2 [Source:HGNC Symbol;Acc:HGNC:9288] | 5792 | 0.162 | 0.0224 | No |
| 19 | C5orf51 | chromosome 5 open reading frame 51 [Source:HGNC Symbol;Acc:HGNC:27750] | 6032 | 0.088 | 0.0169 | No |
| 20 | ZNF138 | zinc finger protein 138 [Source:HGNC Symbol;Acc:HGNC:12922] | 9410 | 0.000 | -0.0743 | No |
| 21 | NEFL | neurofilament light [Source:HGNC Symbol;Acc:HGNC:7739] | 9722 | 0.000 | -0.0827 | No |
| 22 | KCNT2 | potassium sodium-activated channel subfamily T member 2 [Source:HGNC Symbol;Acc:HGNC:18866] | 9933 | 0.000 | -0.0884 | No |
| 23 | SLURP2 | secreted LY6/PLAUR domain containing 2 [Source:HGNC Symbol;Acc:HGNC:25549] | 10285 | 0.000 | -0.0979 | No |
| 24 | STOML3 | stomatin like 3 [Source:HGNC Symbol;Acc:HGNC:19420] | 11411 | 0.000 | -0.1283 | No |
| 25 | NTSR1 | neurotensin receptor 1 [Source:HGNC Symbol;Acc:HGNC:8039] | 11986 | 0.000 | -0.1438 | No |
| 26 | ARL17B | ADP ribosylation factor like GTPase 17B [Source:HGNC Symbol;Acc:HGNC:32387] | 12268 | 0.000 | -0.1514 | No |
| 27 | ARL17A | ADP ribosylation factor like GTPase 17A [Source:HGNC Symbol;Acc:HGNC:24096] | 12271 | 0.000 | -0.1515 | No |
| 28 | IGF2 | insulin like growth factor 2 [Source:HGNC Symbol;Acc:HGNC:5466] | 12475 | 0.000 | -0.1569 | No |
| 29 | DSCAML1 | DS cell adhesion molecule like 1 [Source:HGNC Symbol;Acc:HGNC:14656] | 12944 | 0.000 | -0.1696 | No |
| 30 | GDF2 | growth differentiation factor 2 [Source:HGNC Symbol;Acc:HGNC:4217] | 13447 | 0.000 | -0.1831 | No |
| 31 | FAM218A | family with sequence similarity 218 member A [Source:HGNC Symbol;Acc:HGNC:26466] | 14102 | 0.000 | -0.2008 | No |
| 32 | STUM | stum, mechanosensory transduction mediator homolog [Source:HGNC Symbol;Acc:HGNC:30491] | 15525 | 0.000 | -0.2392 | No |
| 33 | PPP1R3B | protein phosphatase 1 regulatory subunit 3B [Source:HGNC Symbol;Acc:HGNC:14942] | 17657 | 0.000 | -0.2968 | No |
| 34 | SEMA6A | semaphorin 6A [Source:HGNC Symbol;Acc:HGNC:10738] | 17900 | 0.000 | -0.3034 | No |
| 35 | MPL | MPL proto-oncogene, thrombopoietin receptor [Source:HGNC Symbol;Acc:HGNC:7217] | 19109 | 0.000 | -0.3360 | No |
| 36 | IFI44L | interferon induced protein 44 like [Source:HGNC Symbol;Acc:HGNC:17817] | 20820 | 0.000 | -0.3822 | No |
| 37 | CNGB3 | cyclic nucleotide gated channel subunit beta 3 [Source:HGNC Symbol;Acc:HGNC:2153] | 21013 | 0.000 | -0.3874 | No |
| 38 | CPNE5 | copine 5 [Source:HGNC Symbol;Acc:HGNC:2318] | 21397 | 0.000 | -0.3977 | No |
| 39 | TTC22 | tetratricopeptide repeat domain 22 [Source:HGNC Symbol;Acc:HGNC:26067] | 22622 | 0.000 | -0.4308 | No |
| 40 | SLC6A3 | solute carrier family 6 member 3 [Source:HGNC Symbol;Acc:HGNC:11049] | 23432 | 0.000 | -0.4527 | No |
| 41 | C4orf19 | chromosome 4 open reading frame 19 [Source:HGNC Symbol;Acc:HGNC:25618] | 25420 | 0.000 | -0.5064 | No |
| 42 | PCSK1N | proprotein convertase subtilisin/kexin type 1 inhibitor [Source:HGNC Symbol;Acc:HGNC:17301] | 25563 | 0.000 | -0.5102 | No |
| 43 | ADAMTS14 | ADAM metallopeptidase with thrombospondin type 1 motif 14 [Source:HGNC Symbol;Acc:HGNC:14899] | 25671 | 0.000 | -0.5131 | No |
| 44 | C19orf84 | chromosome 19 open reading frame 84 [Source:HGNC Symbol;Acc:HGNC:27112] | 26909 | 0.000 | -0.5465 | No |
| 45 | C9orf50 | chromosome 9 open reading frame 50 [Source:HGNC Symbol;Acc:HGNC:23677] | 28062 | 0.000 | -0.5776 | No |
| 46 | CD1E | CD1e molecule [Source:HGNC Symbol;Acc:HGNC:1638] | 28092 | 0.000 | -0.5784 | No |
| 47 | C2orf88 | chromosome 2 open reading frame 88 [Source:HGNC Symbol;Acc:HGNC:28191] | 29292 | -0.195 | -0.6087 | No |
| 48 | VWF | von Willebrand factor [Source:HGNC Symbol;Acc:HGNC:12726] | 29921 | -0.336 | -0.6220 | No |
| 49 | CYP26B1 | cytochrome P450 family 26 subfamily B member 1 [Source:HGNC Symbol;Acc:HGNC:20581] | 30087 | -0.376 | -0.6224 | No |
| 50 | IDH3A | isocitrate dehydrogenase (NAD(+)) 3 catalytic subunit alpha [Source:HGNC Symbol;Acc:HGNC:5384] | 30345 | -0.445 | -0.6245 | No |
| 51 | RNF2 | ring finger protein 2 [Source:HGNC Symbol;Acc:HGNC:10061] | 30547 | -0.497 | -0.6246 | No |
| 52 | TNFSF8 | TNF superfamily member 8 [Source:HGNC Symbol;Acc:HGNC:11938] | 30658 | -0.528 | -0.6218 | No |
| 53 | PHF12 | PHD finger protein 12 [Source:HGNC Symbol;Acc:HGNC:20816] | 30676 | -0.532 | -0.6165 | No |
| 54 | NHSL2 | NHS like 2 [Source:HGNC Symbol;Acc:HGNC:33737] | 30714 | -0.543 | -0.6116 | No |
| 55 | LIMD2 | LIM domain containing 2 [Source:HGNC Symbol;Acc:HGNC:28142] | 31188 | -0.670 | -0.6172 | No |
| 56 | NCBP3 | nuclear cap binding subunit 3 [Source:HGNC Symbol;Acc:HGNC:24612] | 31696 | -0.800 | -0.6222 | No |
| 57 | CHST14 | carbohydrate sulfotransferase 14 [Source:HGNC Symbol;Acc:HGNC:24464] | 32067 | -0.930 | -0.6221 | No |
| 58 | KAT7 | lysine acetyltransferase 7 [Source:HGNC Symbol;Acc:HGNC:17016] | 32590 | -1.116 | -0.6241 | No |
| 59 | MYLIP | myosin regulatory light chain interacting protein [Source:HGNC Symbol;Acc:HGNC:21155] | 32883 | -1.213 | -0.6189 | No |
| 60 | PIP4P1 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1 [Source:HGNC Symbol;Acc:HGNC:19299] | 33169 | -1.319 | -0.6123 | No |
| 61 | ZBTB20 | zinc finger and BTB domain containing 20 [Source:HGNC Symbol;Acc:HGNC:13503] | 34730 | -1.992 | -0.6329 | Yes |
| 62 | FGD1 | FYVE, RhoGEF and PH domain containing 1 [Source:HGNC Symbol;Acc:HGNC:3663] | 34845 | -2.079 | -0.6134 | Yes |
| 63 | DNMT3B | DNA methyltransferase 3 beta [Source:HGNC Symbol;Acc:HGNC:2979] | 35208 | -2.316 | -0.5981 | Yes |
| 64 | IQGAP3 | IQ motif containing GTPase activating protein 3 [Source:HGNC Symbol;Acc:HGNC:20669] | 35363 | -2.359 | -0.5768 | Yes |
| 65 | GANC | glucosidase alpha, neutral C [Source:HGNC Symbol;Acc:HGNC:4139] | 35623 | -2.585 | -0.5558 | Yes |
| 66 | JADE1 | jade family PHD finger 1 [Source:HGNC Symbol;Acc:HGNC:30027] | 35733 | -2.662 | -0.5299 | Yes |
| 67 | TDP2 | tyrosyl-DNA phosphodiesterase 2 [Source:HGNC Symbol;Acc:HGNC:17768] | 35750 | -2.690 | -0.5012 | Yes |
| 68 | ERF | ETS2 repressor factor [Source:HGNC Symbol;Acc:HGNC:3444] | 35771 | -2.704 | -0.4724 | Yes |
| 69 | MEX3A | mex-3 RNA binding family member A [Source:HGNC Symbol;Acc:HGNC:33482] | 35937 | -2.850 | -0.4460 | Yes |
| 70 | HCFC1R1 | host cell factor C1 regulator 1 [Source:HGNC Symbol;Acc:HGNC:21198] | 36002 | -2.926 | -0.4160 | Yes |
| 71 | ECH1 | enoyl-CoA hydratase 1 [Source:HGNC Symbol;Acc:HGNC:3149] | 36133 | -3.086 | -0.3861 | Yes |
| 72 | PLXNA2 | plexin A2 [Source:HGNC Symbol;Acc:HGNC:9100] | 36137 | -3.089 | -0.3528 | Yes |
| 73 | ARHGAP33 | Rho GTPase activating protein 33 [Source:HGNC Symbol;Acc:HGNC:23085] | 36273 | -3.204 | -0.3217 | Yes |
| 74 | ZBTB4 | zinc finger and BTB domain containing 4 [Source:HGNC Symbol;Acc:HGNC:23847] | 36409 | -3.395 | -0.2886 | Yes |
| 75 | RND2 | Rho family GTPase 2 [Source:HGNC Symbol;Acc:HGNC:18315] | 36442 | -3.442 | -0.2522 | Yes |
| 76 | NECTIN1 | nectin cell adhesion molecule 1 [Source:HGNC Symbol;Acc:HGNC:9706] | 36476 | -3.457 | -0.2156 | Yes |
| 77 | KYAT1 | kynurenine aminotransferase 1 [Source:HGNC Symbol;Acc:HGNC:1564] | 36740 | -3.899 | -0.1805 | Yes |
| 78 | TMEM218 | transmembrane protein 218 [Source:HGNC Symbol;Acc:HGNC:27344] | 36811 | -4.010 | -0.1389 | Yes |
| 79 | FRMD4B | FERM domain containing 4B [Source:HGNC Symbol;Acc:HGNC:24886] | 36927 | -4.302 | -0.0954 | Yes |
| 80 | MTSS1 | MTSS I-BAR domain containing 1 [Source:HGNC Symbol;Acc:HGNC:20443] | 36939 | -4.337 | -0.0488 | Yes |
| 81 | SEMA4G | semaphorin 4G [Source:HGNC Symbol;Acc:HGNC:10735] | 37065 | -4.876 | 0.0007 | Yes |

