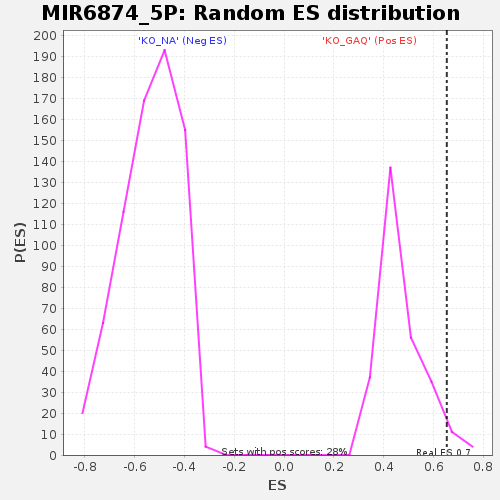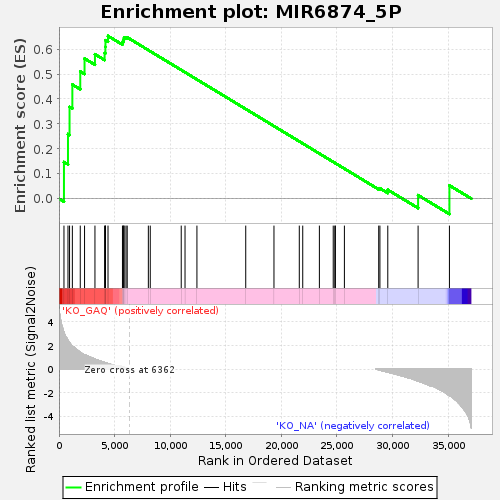
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_GAQ |
| GeneSet | MIR6874_5P |
| Enrichment Score (ES) | 0.65295887 |
| Normalized Enrichment Score (NES) | 1.390544 |
| Nominal p-value | 0.042857144 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SH3PXD2B | SH3 and PX domains 2B [Source:HGNC Symbol;Acc:HGNC:29242] | 446 | 3.125 | 0.1470 | Yes |
| 2 | IFFO2 | intermediate filament family orphan 2 [Source:HGNC Symbol;Acc:HGNC:27006] | 808 | 2.399 | 0.2594 | Yes |
| 3 | KPNA4 | karyopherin subunit alpha 4 [Source:HGNC Symbol;Acc:HGNC:6397] | 949 | 2.214 | 0.3683 | Yes |
| 4 | ARHGEF7 | Rho guanine nucleotide exchange factor 7 [Source:HGNC Symbol;Acc:HGNC:15607] | 1201 | 1.895 | 0.4579 | Yes |
| 5 | SLC1A4 | solute carrier family 1 member 4 [Source:HGNC Symbol;Acc:HGNC:10942] | 1912 | 1.409 | 0.5105 | Yes |
| 6 | SERTAD2 | SERTA domain containing 2 [Source:HGNC Symbol;Acc:HGNC:30784] | 2298 | 1.224 | 0.5624 | Yes |
| 7 | ANAPC16 | anaphase promoting complex subunit 16 [Source:HGNC Symbol;Acc:HGNC:26976] | 3233 | 0.832 | 0.5795 | Yes |
| 8 | DVL3 | dishevelled segment polarity protein 3 [Source:HGNC Symbol;Acc:HGNC:3087] | 4102 | 0.549 | 0.5840 | Yes |
| 9 | HOXA5 | homeobox A5 [Source:HGNC Symbol;Acc:HGNC:5106] | 4171 | 0.528 | 0.6090 | Yes |
| 10 | FYTTD1 | forty-two-three domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25407] | 4178 | 0.526 | 0.6356 | Yes |
| 11 | BFAR | bifunctional apoptosis regulator [Source:HGNC Symbol;Acc:HGNC:17613] | 4408 | 0.462 | 0.6530 | Yes |
| 12 | GALNT7 | polypeptide N-acetylgalactosaminyltransferase 7 [Source:HGNC Symbol;Acc:HGNC:4129] | 5707 | 0.188 | 0.6275 | No |
| 13 | NFS1 | NFS1 cysteine desulfurase [Source:HGNC Symbol;Acc:HGNC:15910] | 5772 | 0.167 | 0.6343 | No |
| 14 | RNF44 | ring finger protein 44 [Source:HGNC Symbol;Acc:HGNC:19180] | 5827 | 0.151 | 0.6405 | No |
| 15 | OSBPL7 | oxysterol binding protein like 7 [Source:HGNC Symbol;Acc:HGNC:16387] | 5855 | 0.144 | 0.6471 | No |
| 16 | CADM2 | cell adhesion molecule 2 [Source:HGNC Symbol;Acc:HGNC:29849] | 6025 | 0.090 | 0.6471 | No |
| 17 | SMAD5 | SMAD family member 5 [Source:HGNC Symbol;Acc:HGNC:6771] | 6134 | 0.060 | 0.6473 | No |
| 18 | NETO1 | neuropilin and tolloid like 1 [Source:HGNC Symbol;Acc:HGNC:13823] | 8036 | 0.000 | 0.5960 | No |
| 19 | CD247 | CD247 molecule [Source:HGNC Symbol;Acc:HGNC:1677] | 8208 | 0.000 | 0.5913 | No |
| 20 | MIXL1 | Mix paired-like homeobox [Source:HGNC Symbol;Acc:HGNC:13363] | 10993 | 0.000 | 0.5162 | No |
| 21 | GFRA2 | GDNF family receptor alpha 2 [Source:HGNC Symbol;Acc:HGNC:4244] | 11335 | 0.000 | 0.5070 | No |
| 22 | SPX | spexin hormone [Source:HGNC Symbol;Acc:HGNC:28139] | 12405 | 0.000 | 0.4782 | No |
| 23 | SYBU | syntabulin [Source:HGNC Symbol;Acc:HGNC:26011] | 16795 | 0.000 | 0.3597 | No |
| 24 | GCNT4 | glucosaminyl (N-acetyl) transferase 4 [Source:HGNC Symbol;Acc:HGNC:17973] | 19334 | 0.000 | 0.2912 | No |
| 25 | FAM155B | family with sequence similarity 155 member B [Source:HGNC Symbol;Acc:HGNC:30701] | 21612 | 0.000 | 0.2298 | No |
| 26 | LSAMP | limbic system associated membrane protein [Source:HGNC Symbol;Acc:HGNC:6705] | 21922 | 0.000 | 0.2214 | No |
| 27 | GIPC3 | GIPC PDZ domain containing family member 3 [Source:HGNC Symbol;Acc:HGNC:18183] | 23419 | 0.000 | 0.1811 | No |
| 28 | DGKI | diacylglycerol kinase iota [Source:HGNC Symbol;Acc:HGNC:2855] | 24664 | 0.000 | 0.1475 | No |
| 29 | TRPC6 | transient receptor potential cation channel subfamily C member 6 [Source:HGNC Symbol;Acc:HGNC:12338] | 24806 | 0.000 | 0.1437 | No |
| 30 | KLHDC8A | kelch domain containing 8A [Source:HGNC Symbol;Acc:HGNC:25573] | 24861 | 0.000 | 0.1422 | No |
| 31 | ADAMTS15 | ADAM metallopeptidase with thrombospondin type 1 motif 15 [Source:HGNC Symbol;Acc:HGNC:16305] | 25669 | 0.000 | 0.1204 | No |
| 32 | RAB3B | RAB3B, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9778] | 28755 | -0.061 | 0.0403 | No |
| 33 | RAB3C | RAB3C, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:30269] | 28864 | -0.088 | 0.0418 | No |
| 34 | MAP4K4 | mitogen-activated protein kinase kinase kinase kinase 4 [Source:HGNC Symbol;Acc:HGNC:6866] | 29571 | -0.255 | 0.0358 | No |
| 35 | NFAT5 | nuclear factor of activated T cells 5 [Source:HGNC Symbol;Acc:HGNC:7774] | 32297 | -1.022 | 0.0143 | No |
| 36 | ZNF503 | zinc finger protein 503 [Source:HGNC Symbol;Acc:HGNC:23589] | 35115 | -2.261 | 0.0533 | No |