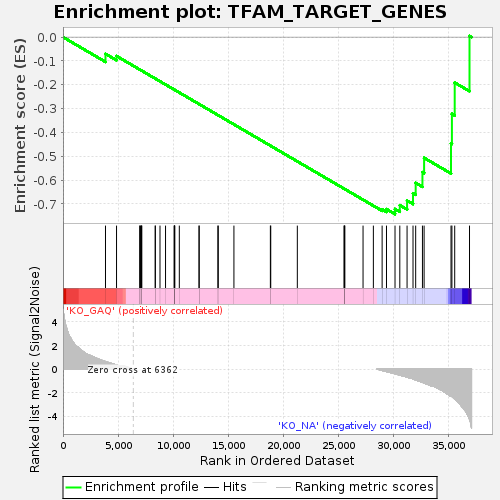
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | TFAM_TARGET_GENES |
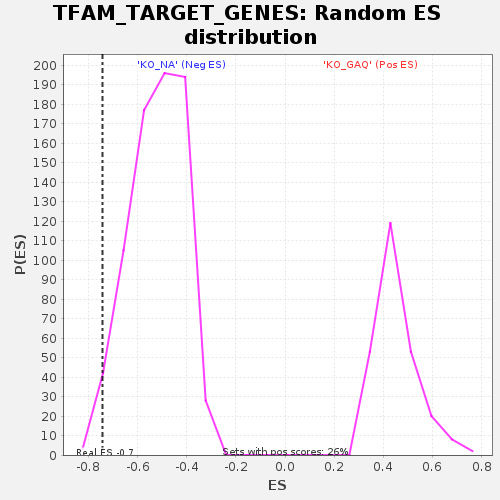
| Enrichment Score (ES) | -0.7429884 |
| Normalized Enrichment Score (NES) | -1.4253734 |
| Nominal p-value | 0.020134227 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MTCO1P12 | MT-CO1 pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:52014] | 3857 | 0.621 | -0.0702 | No |
| 2 | MTCO3P22 | MT-CO3 pseudogene 22 [Source:HGNC Symbol;Acc:HGNC:52125] | 4860 | 0.326 | -0.0794 | No |
| 3 | MTND2P4 | MT-ND2 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:42105] | 6976 | 0.000 | -0.1365 | No |
| 4 | MTND2P8 | MT-ND2 pseudogene 8 [Source:HGNC Symbol;Acc:HGNC:42109] | 6981 | 0.000 | -0.1366 | No |
| 5 | MTCO1P2 | MT-CO1 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:19932] | 7032 | 0.000 | -0.1380 | No |
| 6 | MTND1P7 | MT-ND1 pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:42102] | 7136 | 0.000 | -0.1408 | No |
| 7 | MTND1P6 | MT-ND1 pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:42103] | 7137 | 0.000 | -0.1408 | No |
| 8 | MTND1P9 | MT-ND1 pseudogene 9 [Source:HGNC Symbol;Acc:HGNC:42099] | 7138 | 0.000 | -0.1408 | No |
| 9 | MTATP6P3 | MT-ATP6 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:44577] | 8376 | 0.000 | -0.1742 | No |
| 10 | MTATP6P2 | MT-ATP6 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:44576] | 8377 | 0.000 | -0.1742 | No |
| 11 | MTND4LP1 | MT-ND4L pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:31347] | 8805 | 0.000 | -0.1857 | No |
| 12 | NMTRS-TGA3-1 | nuclear-encoded mitochondrial tRNA-Ser (TGA) 3-1 [Source:HGNC Symbol;Acc:HGNC:35087] | 9308 | 0.000 | -0.1992 | No |
| 13 | MTND1P11 | MT-ND1 pseudogene 11 [Source:HGNC Symbol;Acc:HGNC:42060] | 10101 | 0.000 | -0.2206 | No |
| 14 | MTND1P14 | MT-ND1 pseudogene 14 [Source:HGNC Symbol;Acc:HGNC:42063] | 10102 | 0.000 | -0.2206 | No |
| 15 | MTND1P36 | MT-ND1 pseudogene 36 [Source:HGNC Symbol;Acc:HGNC:42086] | 10117 | 0.000 | -0.2210 | No |
| 16 | MTND1P22 | MT-ND1 pseudogene 22 [Source:HGNC Symbol;Acc:HGNC:42093] | 10134 | 0.000 | -0.2214 | No |
| 17 | MTND1P15 | MT-ND1 pseudogene 15 [Source:HGNC Symbol;Acc:HGNC:42064] | 10143 | 0.000 | -0.2216 | No |
| 18 | MTND1P37 | MT-ND1 pseudogene 37 [Source:HGNC Symbol;Acc:HGNC:52314] | 10559 | 0.000 | -0.2328 | No |
| 19 | MTRNR2L8 | MT-RNR2 like 8 [Source:HGNC Symbol;Acc:HGNC:37165] | 12353 | 0.000 | -0.2812 | No |
| 20 | MTRNR2L9 | MT-RNR2 like 9 [Source:HGNC Symbol;Acc:HGNC:37166] | 12356 | 0.000 | -0.2813 | No |
| 21 | MTRNR2L12 | MT-RNR2 like 12 [Source:HGNC Symbol;Acc:HGNC:37169] | 14060 | 0.000 | -0.3273 | No |
| 22 | MTCO3P12 | MT-CO3 pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:52042] | 14093 | 0.000 | -0.3281 | No |
| 23 | RNU6-656P | RNA, U6 small nuclear 656, pseudogene [Source:HGNC Symbol;Acc:HGNC:47619] | 15516 | 0.000 | -0.3665 | No |
| 24 | MTND4P24 | MT-ND4 pseudogene 24 [Source:HGNC Symbol;Acc:HGNC:42220] | 18825 | 0.000 | -0.4558 | No |
| 25 | MTND4P12 | MT-ND4 pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:42199] | 18859 | 0.000 | -0.4567 | No |
| 26 | MTND6P4 | MT-ND6 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:39467] | 21272 | 0.000 | -0.5218 | No |
| 27 | MTND2P24 | MT-ND2 pseudogene 24 [Source:HGNC Symbol;Acc:HGNC:42125] | 25520 | 0.000 | -0.6364 | No |
| 28 | MTND2P16 | MT-ND2 pseudogene 16 [Source:HGNC Symbol;Acc:HGNC:42117] | 25532 | 0.000 | -0.6367 | No |
| 29 | MTND2P38 | MT-ND2 pseudogene 38 [Source:HGNC Symbol;Acc:HGNC:52069] | 25566 | 0.000 | -0.6376 | No |
| 30 | MTND2P33 | MT-ND2 pseudogene 33 [Source:HGNC Symbol;Acc:HGNC:42134] | 25578 | 0.000 | -0.6379 | No |
| 31 | AL669831.3 | septin 14 (SEPT14) pseudogene | 27238 | 0.000 | -0.6827 | No |
| 32 | MTND4LP19 | MT-ND4L pseudogene 19 [Source:HGNC Symbol;Acc:HGNC:42253] | 28175 | 0.000 | -0.7080 | No |
| 33 | MTND5P11 | MT-ND5 pseudogene 11 [Source:HGNC Symbol;Acc:HGNC:42273] | 28970 | -0.114 | -0.7232 | Yes |
| 34 | MTND5P10 | MT-ND5 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:42272] | 29366 | -0.215 | -0.7221 | Yes |
| 35 | MTCYBP35 | MT-CYB pseudogene 35 [Source:HGNC Symbol;Acc:HGNC:52303] | 30140 | -0.392 | -0.7216 | Yes |
| 36 | MTCO1P45 | MT-CO1 pseudogene 45 [Source:HGNC Symbol;Acc:HGNC:52110] | 30575 | -0.506 | -0.7057 | Yes |
| 37 | MTND1P30 | MT-ND1 pseudogene 30 [Source:HGNC Symbol;Acc:HGNC:42079] | 31234 | -0.682 | -0.6862 | Yes |
| 38 | MTCO3P13 | MT-CO3 pseudogene 13 [Source:HGNC Symbol;Acc:HGNC:52043] | 31774 | -0.830 | -0.6555 | Yes |
| 39 | MTND3P19 | MT-ND3 pseudogene 19 [Source:HGNC Symbol;Acc:HGNC:52081] | 32016 | -0.916 | -0.6120 | Yes |
| 40 | MTCO1P25 | MT-CO1 pseudogene 25 [Source:HGNC Symbol;Acc:HGNC:52090] | 32630 | -1.128 | -0.5670 | Yes |
| 41 | RPL18AP8 | ribosomal protein L18a pseudogene 8 [Source:HGNC Symbol;Acc:HGNC:36984] | 32781 | -1.177 | -0.5068 | Yes |
| 42 | MTRNR2L1 | MT-RNR2 like 1 [Source:HGNC Symbol;Acc:HGNC:37155] | 35223 | -2.316 | -0.4463 | Yes |
| 43 | MTCYBP18 | MT-CYB pseudogene 18 [Source:HGNC Symbol;Acc:HGNC:51976] | 35305 | -2.318 | -0.3220 | Yes |
| 44 | SLC39A10 | solute carrier family 39 member 10 [Source:HGNC Symbol;Acc:HGNC:20861] | 35559 | -2.526 | -0.1910 | Yes |
| 45 | ARHGAP15 | Rho GTPase activating protein 15 [Source:HGNC Symbol;Acc:HGNC:21030] | 36903 | -4.256 | 0.0050 | Yes |