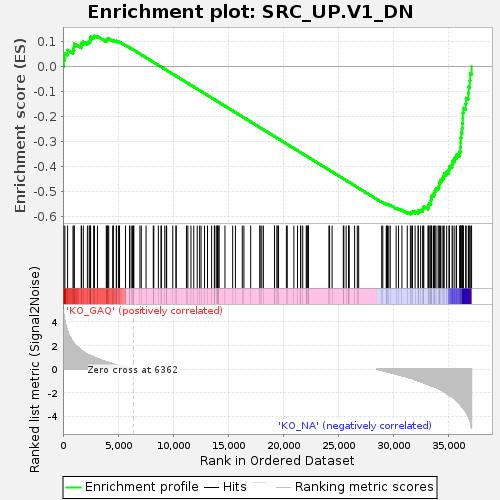
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | SRC_UP.V1_DN |
| Enrichment Score (ES) | -0.5900125 |
| Normalized Enrichment Score (NES) | -1.3089466 |
| Nominal p-value | 0.02 |
| FDR q-value | 0.17624381 |
| FWER p-Value | 0.978 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SERPINB2 | serpin family B member 2 [Source:HGNC Symbol;Acc:HGNC:8584] | 74 | 4.552 | 0.0274 | No |
| 2 | IFIT1 | interferon induced protein with tetratricopeptide repeats 1 [Source:HGNC Symbol;Acc:HGNC:5407] | 170 | 4.018 | 0.0507 | No |
| 3 | GALNTL6 | polypeptide N-acetylgalactosaminyltransferase like 6 [Source:HGNC Symbol;Acc:HGNC:33844] | 416 | 3.149 | 0.0644 | No |
| 4 | ACSL4 | acyl-CoA synthetase long chain family member 4 [Source:HGNC Symbol;Acc:HGNC:3571] | 907 | 2.278 | 0.0658 | No |
| 5 | ZNF436 | zinc finger protein 436 [Source:HGNC Symbol;Acc:HGNC:20814] | 999 | 2.155 | 0.0773 | No |
| 6 | WASH3P | WASP family homolog 3, pseudogene [Source:HGNC Symbol;Acc:HGNC:24362] | 1021 | 2.123 | 0.0904 | No |
| 7 | POP1 | POP1 homolog, ribonuclease P/MRP subunit [Source:HGNC Symbol;Acc:HGNC:30129] | 1652 | 1.590 | 0.0836 | No |
| 8 | RRN3 | RRN3 homolog, RNA polymerase I transcription factor [Source:HGNC Symbol;Acc:HGNC:30346] | 1682 | 1.572 | 0.0930 | No |
| 9 | COP1 | COP1 E3 ubiquitin ligase [Source:HGNC Symbol;Acc:HGNC:17440] | 1844 | 1.467 | 0.0981 | No |
| 10 | HS3ST3A1 | heparan sulfate-glucosamine 3-sulfotransferase 3A1 [Source:HGNC Symbol;Acc:HGNC:5196] | 2213 | 1.230 | 0.0961 | No |
| 11 | STARD3 | StAR related lipid transfer domain containing 3 [Source:HGNC Symbol;Acc:HGNC:17579] | 2374 | 1.179 | 0.0993 | No |
| 12 | COMMD9 | COMM domain containing 9 [Source:HGNC Symbol;Acc:HGNC:25014] | 2464 | 1.141 | 0.1043 | No |
| 13 | RAB33B | RAB33B, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:16075] | 2475 | 1.138 | 0.1114 | No |
| 14 | COQ4 | coenzyme Q4 [Source:HGNC Symbol;Acc:HGNC:19693] | 2506 | 1.127 | 0.1178 | No |
| 15 | IRX5 | iroquois homeobox 5 [Source:HGNC Symbol;Acc:HGNC:14361] | 2784 | 1.014 | 0.1169 | No |
| 16 | DTWD1 | DTW domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30926] | 2854 | 0.982 | 0.1213 | No |
| 17 | ABHD3 | abhydrolase domain containing 3, phospholipase [Source:HGNC Symbol;Acc:HGNC:18718] | 3130 | 0.868 | 0.1195 | No |
| 18 | STK17B | serine/threonine kinase 17b [Source:HGNC Symbol;Acc:HGNC:11396] | 3928 | 0.598 | 0.1018 | No |
| 19 | CMTR2 | cap methyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:25635] | 3978 | 0.586 | 0.1042 | No |
| 20 | OTUD6B | OTU deubiquitinase 6B [Source:HGNC Symbol;Acc:HGNC:24281] | 4020 | 0.572 | 0.1068 | No |
| 21 | ZNF451 | zinc finger protein 451 [Source:HGNC Symbol;Acc:HGNC:21091] | 4075 | 0.557 | 0.1089 | No |
| 22 | ZNF3 | zinc finger protein 3 [Source:HGNC Symbol;Acc:HGNC:13089] | 4153 | 0.532 | 0.1103 | No |
| 23 | POLR1A | RNA polymerase I subunit A [Source:HGNC Symbol;Acc:HGNC:17264] | 4511 | 0.427 | 0.1034 | No |
| 24 | ZNF628 | zinc finger protein 628 [Source:HGNC Symbol;Acc:HGNC:28054] | 4574 | 0.411 | 0.1043 | No |
| 25 | MYSM1 | Myb like, SWIRM and MPN domains 1 [Source:HGNC Symbol;Acc:HGNC:29401] | 4837 | 0.334 | 0.0994 | No |
| 26 | CSAD | cysteine sulfinic acid decarboxylase [Source:HGNC Symbol;Acc:HGNC:18966] | 4866 | 0.325 | 0.1007 | No |
| 27 | RTN4IP1 | reticulon 4 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:18647] | 5045 | 0.274 | 0.0977 | No |
| 28 | PTGER4 | prostaglandin E receptor 4 [Source:HGNC Symbol;Acc:HGNC:9596] | 5132 | 0.248 | 0.0970 | No |
| 29 | NOL12 | nucleolar protein 12 [Source:HGNC Symbol;Acc:HGNC:28585] | 5673 | 0.198 | 0.0836 | No |
| 30 | KIAA1841 | KIAA1841 [Source:HGNC Symbol;Acc:HGNC:29387] | 6052 | 0.083 | 0.0739 | No |
| 31 | MON1A | MON1 homolog A, secretory trafficking associated [Source:HGNC Symbol;Acc:HGNC:28207] | 6062 | 0.081 | 0.0742 | No |
| 32 | RNF168 | ring finger protein 168 [Source:HGNC Symbol;Acc:HGNC:26661] | 6251 | 0.028 | 0.0693 | No |
| 33 | PPIL1 | peptidylprolyl isomerase like 1 [Source:HGNC Symbol;Acc:HGNC:9260] | 6302 | 0.013 | 0.0680 | No |
| 34 | C11orf72 | chromosome 11 putative open reading frame 72 [Source:HGNC Symbol;Acc:HGNC:26915] | 6403 | 0.000 | 0.0653 | No |
| 35 | PRSS33 | serine protease 33 [Source:HGNC Symbol;Acc:HGNC:30405] | 6431 | 0.000 | 0.0646 | No |
| 36 | CWH43 | cell wall biogenesis 43 C-terminal homolog [Source:HGNC Symbol;Acc:HGNC:26133] | 6985 | 0.000 | 0.0496 | No |
| 37 | DGCR11 | DiGeorge syndrome critical region gene 11 [Source:HGNC Symbol;Acc:HGNC:17226] | 7105 | 0.000 | 0.0464 | No |
| 38 | ARSJ | arylsulfatase family member J [Source:HGNC Symbol;Acc:HGNC:26286] | 7539 | 0.000 | 0.0346 | No |
| 39 | DEPP1 | DEPP1 autophagy regulator [Source:HGNC Symbol;Acc:HGNC:23355] | 8195 | 0.000 | 0.0169 | No |
| 40 | WT1-AS | WT1 antisense RNA [Source:HGNC Symbol;Acc:HGNC:18135] | 8244 | 0.000 | 0.0156 | No |
| 41 | S100A9 | S100 calcium binding protein A9 [Source:HGNC Symbol;Acc:HGNC:10499] | 8656 | 0.000 | 0.0045 | No |
| 42 | SH3TC2 | SH3 domain and tetratricopeptide repeats 2 [Source:HGNC Symbol;Acc:HGNC:29427] | 8896 | 0.000 | -0.0020 | No |
| 43 | PRR19 | proline rich 19 [Source:HGNC Symbol;Acc:HGNC:33728] | 8932 | 0.000 | -0.0030 | No |
| 44 | NPY5R | neuropeptide Y receptor Y5 [Source:HGNC Symbol;Acc:HGNC:7958] | 9240 | 0.000 | -0.0113 | No |
| 45 | GRID1 | glutamate ionotropic receptor delta type subunit 1 [Source:HGNC Symbol;Acc:HGNC:4575] | 9376 | 0.000 | -0.0149 | No |
| 46 | ZNF135 | zinc finger protein 135 [Source:HGNC Symbol;Acc:HGNC:12919] | 9411 | 0.000 | -0.0158 | No |
| 47 | PLAT | plasminogen activator, tissue type [Source:HGNC Symbol;Acc:HGNC:9051] | 9965 | 0.000 | -0.0308 | No |
| 48 | SPRR1B | small proline rich protein 1B [Source:HGNC Symbol;Acc:HGNC:11260] | 10254 | 0.000 | -0.0386 | No |
| 49 | MEGF6 | multiple EGF like domains 6 [Source:HGNC Symbol;Acc:HGNC:3232] | 10275 | 0.000 | -0.0392 | No |
| 50 | ADH4 | alcohol dehydrogenase 4 (class II), pi polypeptide [Source:HGNC Symbol;Acc:HGNC:252] | 11208 | 0.000 | -0.0644 | No |
| 51 | GFRA3 | GDNF family receptor alpha 3 [Source:HGNC Symbol;Acc:HGNC:4245] | 11334 | 0.000 | -0.0678 | No |
| 52 | EREG | epiregulin [Source:HGNC Symbol;Acc:HGNC:3443] | 11617 | 0.000 | -0.0754 | No |
| 53 | C17orf64 | chromosome 17 open reading frame 64 [Source:HGNC Symbol;Acc:HGNC:26990] | 11870 | 0.000 | -0.0823 | No |
| 54 | LYPD3 | LY6/PLAUR domain containing 3 [Source:HGNC Symbol;Acc:HGNC:24880] | 12184 | 0.000 | -0.0907 | No |
| 55 | SPDYE1 | speedy/RINGO cell cycle regulator family member E1 [Source:HGNC Symbol;Acc:HGNC:16408] | 12404 | 0.000 | -0.0967 | No |
| 56 | TGM5 | transglutaminase 5 [Source:HGNC Symbol;Acc:HGNC:11781] | 12531 | 0.000 | -0.1001 | No |
| 57 | CAPN12 | calpain 12 [Source:HGNC Symbol;Acc:HGNC:13249] | 12849 | 0.000 | -0.1087 | No |
| 58 | ZBED2 | zinc finger BED-type containing 2 [Source:HGNC Symbol;Acc:HGNC:20710] | 13116 | 0.000 | -0.1159 | No |
| 59 | TMEM14A | transmembrane protein 14A [Source:HGNC Symbol;Acc:HGNC:21076] | 13492 | 0.000 | -0.1260 | No |
| 60 | SYT15 | synaptotagmin 15 [Source:HGNC Symbol;Acc:HGNC:17167] | 13740 | 0.000 | -0.1327 | No |
| 61 | MGARP | mitochondria localized glutamic acid rich protein [Source:HGNC Symbol;Acc:HGNC:29969] | 13786 | 0.000 | -0.1339 | No |
| 62 | IL7R | interleukin 7 receptor [Source:HGNC Symbol;Acc:HGNC:6024] | 13957 | 0.000 | -0.1385 | No |
| 63 | NYNRIN | NYN domain and retroviral integrase containing [Source:HGNC Symbol;Acc:HGNC:20165] | 14012 | 0.000 | -0.1400 | No |
| 64 | PCDHB4 | protocadherin beta 4 [Source:HGNC Symbol;Acc:HGNC:8689] | 14031 | 0.000 | -0.1405 | No |
| 65 | PIK3C2G | phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma [Source:HGNC Symbol;Acc:HGNC:8973] | 14155 | 0.000 | -0.1438 | No |
| 66 | PIK3C2B | phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta [Source:HGNC Symbol;Acc:HGNC:8972] | 14157 | 0.000 | -0.1438 | No |
| 67 | PCDH19 | protocadherin 19 [Source:HGNC Symbol;Acc:HGNC:14270] | 14703 | 0.000 | -0.1586 | No |
| 68 | TRIM73 | tripartite motif containing 73 [Source:HGNC Symbol;Acc:HGNC:18162] | 15404 | 0.000 | -0.1776 | No |
| 69 | C8B | complement C8 beta chain [Source:HGNC Symbol;Acc:HGNC:1353] | 15654 | 0.000 | -0.1843 | No |
| 70 | SLC7A14 | solute carrier family 7 member 14 [Source:HGNC Symbol;Acc:HGNC:29326] | 16276 | 0.000 | -0.2011 | No |
| 71 | EMB | embigin [Source:HGNC Symbol;Acc:HGNC:30465] | 16421 | 0.000 | -0.2050 | No |
| 72 | SYT1 | synaptotagmin 1 [Source:HGNC Symbol;Acc:HGNC:11509] | 17038 | 0.000 | -0.2217 | No |
| 73 | IVL | involucrin [Source:HGNC Symbol;Acc:HGNC:6187] | 17874 | 0.000 | -0.2443 | No |
| 74 | GTF2H2 | general transcription factor IIH subunit 2 [Source:HGNC Symbol;Acc:HGNC:4656] | 17886 | 0.000 | -0.2446 | No |
| 75 | S1PR3 | sphingosine-1-phosphate receptor 3 [Source:HGNC Symbol;Acc:HGNC:3167] | 18047 | 0.000 | -0.2490 | No |
| 76 | DPYD | dihydropyrimidine dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3012] | 18189 | 0.000 | -0.2528 | No |
| 77 | MMP1 | matrix metallopeptidase 1 [Source:HGNC Symbol;Acc:HGNC:7155] | 19199 | 0.000 | -0.2801 | No |
| 78 | MMP3 | matrix metallopeptidase 3 [Source:HGNC Symbol;Acc:HGNC:7173] | 19203 | 0.000 | -0.2802 | No |
| 79 | COL8A1 | collagen type VIII alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:2215] | 19412 | 0.000 | -0.2858 | No |
| 80 | SPNS3 | sphingolipid transporter 3 (putative) [Source:HGNC Symbol;Acc:HGNC:28433] | 19506 | 0.000 | -0.2883 | No |
| 81 | CXCL5 | C-X-C motif chemokine ligand 5 [Source:HGNC Symbol;Acc:HGNC:10642] | 19567 | 0.000 | -0.2900 | No |
| 82 | LINC00842 | long intergenic non-protein coding RNA 842 [Source:HGNC Symbol;Acc:HGNC:44989] | 20279 | 0.000 | -0.3092 | No |
| 83 | COBL | cordon-bleu WH2 repeat protein [Source:HGNC Symbol;Acc:HGNC:22199] | 20356 | 0.000 | -0.3113 | No |
| 84 | DISP2 | dispatched RND transporter family member 2 [Source:HGNC Symbol;Acc:HGNC:19712] | 20958 | 0.000 | -0.3275 | No |
| 85 | CENPH | centromere protein H [Source:HGNC Symbol;Acc:HGNC:17268] | 21286 | 0.000 | -0.3364 | No |
| 86 | ADGRF1 | adhesion G protein-coupled receptor F1 [Source:HGNC Symbol;Acc:HGNC:18990] | 21572 | 0.000 | -0.3441 | No |
| 87 | FAM153A | family with sequence similarity 153 member A [Source:HGNC Symbol;Acc:HGNC:29940] | 21578 | 0.000 | -0.3443 | No |
| 88 | CLCA4 | chloride channel accessory 4 [Source:HGNC Symbol;Acc:HGNC:2018] | 21763 | 0.000 | -0.3492 | No |
| 89 | KCNMB3 | potassium calcium-activated channel subfamily M regulatory beta subunit 3 [Source:HGNC Symbol;Acc:HGNC:6287] | 22080 | 0.000 | -0.3578 | No |
| 90 | TMCO2 | transmembrane and coiled-coil domains 2 [Source:HGNC Symbol;Acc:HGNC:23312] | 22130 | 0.000 | -0.3591 | No |
| 91 | ANHX | anomalous homeobox [Source:HGNC Symbol;Acc:HGNC:40024] | 22223 | 0.000 | -0.3616 | No |
| 92 | IFITM10 | interferon induced transmembrane protein 10 [Source:HGNC Symbol;Acc:HGNC:40022] | 22251 | 0.000 | -0.3623 | No |
| 93 | ANO7 | anoctamin 7 [Source:HGNC Symbol;Acc:HGNC:31677] | 22275 | 0.000 | -0.3630 | No |
| 94 | ATPSCKMT | ATP synthase c subunit lysine N-methyltransferase [Source:HGNC Symbol;Acc:HGNC:27029] | 24141 | 0.000 | -0.4135 | No |
| 95 | NLRP8 | NLR family pyrin domain containing 8 [Source:HGNC Symbol;Acc:HGNC:22940] | 24199 | 0.000 | -0.4150 | No |
| 96 | ST8SIA2 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:10870] | 24427 | 0.000 | -0.4212 | No |
| 97 | UTS2R | urotensin 2 receptor [Source:HGNC Symbol;Acc:HGNC:4468] | 25446 | 0.000 | -0.4487 | No |
| 98 | IFI6 | interferon alpha inducible protein 6 [Source:HGNC Symbol;Acc:HGNC:4054] | 25495 | 0.000 | -0.4500 | No |
| 99 | MMP10 | matrix metallopeptidase 10 [Source:HGNC Symbol;Acc:HGNC:7156] | 25703 | 0.000 | -0.4557 | No |
| 100 | GJB6 | gap junction protein beta 6 [Source:HGNC Symbol;Acc:HGNC:4288] | 25917 | 0.000 | -0.4614 | No |
| 101 | ZNF789 | zinc finger protein 789 [Source:HGNC Symbol;Acc:HGNC:27801] | 25960 | 0.000 | -0.4626 | No |
| 102 | ZNF782 | zinc finger protein 782 [Source:HGNC Symbol;Acc:HGNC:33110] | 25963 | 0.000 | -0.4626 | No |
| 103 | ZNF721 | zinc finger protein 721 [Source:HGNC Symbol;Acc:HGNC:29425] | 26474 | 0.000 | -0.4764 | No |
| 104 | IRGC | immunity related GTPase cinema [Source:HGNC Symbol;Acc:HGNC:28835] | 26718 | 0.000 | -0.4830 | No |
| 105 | PHLDA2 | pleckstrin homology like domain family A member 2 [Source:HGNC Symbol;Acc:HGNC:12385] | 26841 | 0.000 | -0.4863 | No |
| 106 | CTDP1 | CTD phosphatase subunit 1 [Source:HGNC Symbol;Acc:HGNC:2498] | 28931 | -0.106 | -0.5422 | No |
| 107 | PAK3 | p21 (RAC1) activated kinase 3 [Source:HGNC Symbol;Acc:HGNC:8592] | 29031 | -0.132 | -0.5440 | No |
| 108 | FANCL | FA complementation group L [Source:HGNC Symbol;Acc:HGNC:20748] | 29349 | -0.209 | -0.5513 | No |
| 109 | SEMA6B | semaphorin 6B [Source:HGNC Symbol;Acc:HGNC:10739] | 29359 | -0.213 | -0.5501 | No |
| 110 | DCAF17 | DDB1 and CUL4 associated factor 17 [Source:HGNC Symbol;Acc:HGNC:25784] | 29457 | -0.229 | -0.5513 | No |
| 111 | ACTL6A | actin like 6A [Source:HGNC Symbol;Acc:HGNC:24124] | 29576 | -0.256 | -0.5528 | No |
| 112 | ADGRE1 | adhesion G protein-coupled receptor E1 [Source:HGNC Symbol;Acc:HGNC:3336] | 29705 | -0.292 | -0.5544 | No |
| 113 | SLC12A5 | solute carrier family 12 member 5 [Source:HGNC Symbol;Acc:HGNC:13818] | 30244 | -0.417 | -0.5663 | No |
| 114 | GON7 | GON7 subunit of KEOPS complex [Source:HGNC Symbol;Acc:HGNC:20356] | 30460 | -0.479 | -0.5690 | No |
| 115 | RARA | retinoic acid receptor alpha [Source:HGNC Symbol;Acc:HGNC:9864] | 30767 | -0.558 | -0.5737 | No |
| 116 | CHAC2 | ChaC glutathione specific gamma-glutamylcyclotransferase 2 [Source:HGNC Symbol;Acc:HGNC:32363] | 31253 | -0.689 | -0.5824 | No |
| 117 | NFKBIE | NFKB inhibitor epsilon [Source:HGNC Symbol;Acc:HGNC:7799] | 31535 | -0.768 | -0.5851 | Yes |
| 118 | PTBP2 | polypyrimidine tract binding protein 2 [Source:HGNC Symbol;Acc:HGNC:17662] | 31661 | -0.787 | -0.5834 | Yes |
| 119 | IRAK2 | interleukin 1 receptor associated kinase 2 [Source:HGNC Symbol;Acc:HGNC:6113] | 31728 | -0.813 | -0.5799 | Yes |
| 120 | CARNMT1 | carnosine N-methyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:23435] | 31965 | -0.901 | -0.5805 | Yes |
| 121 | CDK11B | cyclin dependent kinase 11B [Source:HGNC Symbol;Acc:HGNC:1729] | 32242 | -1.002 | -0.5815 | Yes |
| 122 | TTC14 | tetratricopeptide repeat domain 14 [Source:HGNC Symbol;Acc:HGNC:24697] | 32246 | -1.003 | -0.5751 | Yes |
| 123 | SOAT1 | sterol O-acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:11177] | 32466 | -1.069 | -0.5741 | Yes |
| 124 | SNX32 | sorting nexin 32 [Source:HGNC Symbol;Acc:HGNC:26423] | 32660 | -1.135 | -0.5720 | Yes |
| 125 | MIR17HG | miR-17-92a-1 cluster host gene [Source:HGNC Symbol;Acc:HGNC:23564] | 32685 | -1.140 | -0.5653 | Yes |
| 126 | CA7 | carbonic anhydrase 7 [Source:HGNC Symbol;Acc:HGNC:1381] | 32752 | -1.165 | -0.5596 | Yes |
| 127 | SLC2A6 | solute carrier family 2 member 6 [Source:HGNC Symbol;Acc:HGNC:11011] | 33143 | -1.308 | -0.5617 | Yes |
| 128 | RAP1A | RAP1A, member of RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9855] | 33156 | -1.313 | -0.5536 | Yes |
| 129 | HOXB2 | homeobox B2 [Source:HGNC Symbol;Acc:HGNC:5113] | 33246 | -1.356 | -0.5472 | Yes |
| 130 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 33400 | -1.422 | -0.5422 | Yes |
| 131 | ADA | adenosine deaminase [Source:HGNC Symbol;Acc:HGNC:186] | 33402 | -1.423 | -0.5330 | Yes |
| 132 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 33442 | -1.442 | -0.5248 | Yes |
| 133 | MN1 | MN1 proto-oncogene, transcriptional regulator [Source:HGNC Symbol;Acc:HGNC:7180] | 33477 | -1.451 | -0.5163 | Yes |
| 134 | EIF3CL | eukaryotic translation initiation factor 3 subunit C like [Source:HGNC Symbol;Acc:HGNC:26347] | 33634 | -1.452 | -0.5112 | Yes |
| 135 | RNF220 | ring finger protein 220 [Source:HGNC Symbol;Acc:HGNC:25552] | 33740 | -1.490 | -0.5044 | Yes |
| 136 | CDCA7 | cell division cycle associated 7 [Source:HGNC Symbol;Acc:HGNC:14628] | 33754 | -1.495 | -0.4951 | Yes |
| 137 | ARL15 | ADP ribosylation factor like GTPase 15 [Source:HGNC Symbol;Acc:HGNC:25945] | 33861 | -1.542 | -0.4880 | Yes |
| 138 | NBEAL1 | neurobeachin like 1 [Source:HGNC Symbol;Acc:HGNC:20681] | 34053 | -1.643 | -0.4826 | Yes |
| 139 | TMOD1 | tropomodulin 1 [Source:HGNC Symbol;Acc:HGNC:11871] | 34145 | -1.687 | -0.4742 | Yes |
| 140 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34155 | -1.691 | -0.4635 | Yes |
| 141 | SUV39H2 | suppressor of variegation 3-9 homolog 2 [Source:HGNC Symbol;Acc:HGNC:17287] | 34265 | -1.749 | -0.4552 | Yes |
| 142 | CPPED1 | calcineurin like phosphoesterase domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25632] | 34411 | -1.812 | -0.4474 | Yes |
| 143 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 34533 | -1.882 | -0.4385 | Yes |
| 144 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit [Source:HGNC Symbol;Acc:HGNC:3527] | 34602 | -1.924 | -0.4280 | Yes |
| 145 | TREML1 | triggering receptor expressed on myeloid cells like 1 [Source:HGNC Symbol;Acc:HGNC:20434] | 34830 | -2.071 | -0.4208 | Yes |
| 146 | MCM5 | minichromosome maintenance complex component 5 [Source:HGNC Symbol;Acc:HGNC:6948] | 35037 | -2.200 | -0.4121 | Yes |
| 147 | ATAD2 | ATPase family AAA domain containing 2 [Source:HGNC Symbol;Acc:HGNC:30123] | 35076 | -2.228 | -0.3988 | Yes |
| 148 | MAP3K6 | mitogen-activated protein kinase kinase kinase 6 [Source:HGNC Symbol;Acc:HGNC:6858] | 35322 | -2.326 | -0.3904 | Yes |
| 149 | RCBTB2 | RCC1 and BTB domain containing protein 2 [Source:HGNC Symbol;Acc:HGNC:1914] | 35393 | -2.388 | -0.3769 | Yes |
| 150 | LFNG | LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase [Source:HGNC Symbol;Acc:HGNC:6560] | 35571 | -2.538 | -0.3653 | Yes |
| 151 | RACGAP1 | Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] | 35725 | -2.658 | -0.3523 | Yes |
| 152 | GDPD3 | glycerophosphodiester phosphodiesterase domain containing 3 [Source:HGNC Symbol;Acc:HGNC:28638] | 36003 | -2.926 | -0.3409 | Yes |
| 153 | RFTN1 | raftlin, lipid raft linker 1 [Source:HGNC Symbol;Acc:HGNC:30278] | 36086 | -3.014 | -0.3237 | Yes |
| 154 | NRP1 | neuropilin 1 [Source:HGNC Symbol;Acc:HGNC:8004] | 36095 | -3.029 | -0.3043 | Yes |
| 155 | UHRF1 | ubiquitin like with PHD and ring finger domains 1 [Source:HGNC Symbol;Acc:HGNC:12556] | 36104 | -3.045 | -0.2849 | Yes |
| 156 | UNC119 | unc-119 lipid binding chaperone [Source:HGNC Symbol;Acc:HGNC:12565] | 36148 | -3.095 | -0.2661 | Yes |
| 157 | B3GNT3 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 [Source:HGNC Symbol;Acc:HGNC:13528] | 36228 | -3.196 | -0.2476 | Yes |
| 158 | C1orf159 | chromosome 1 open reading frame 159 [Source:HGNC Symbol;Acc:HGNC:26062] | 36259 | -3.196 | -0.2278 | Yes |
| 159 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36290 | -3.231 | -0.2077 | Yes |
| 160 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36293 | -3.235 | -0.1869 | Yes |
| 161 | PC | pyruvate carboxylase [Source:HGNC Symbol;Acc:HGNC:8636] | 36347 | -3.313 | -0.1670 | Yes |
| 162 | ARRDC3 | arrestin domain containing 3 [Source:HGNC Symbol;Acc:HGNC:29263] | 36557 | -3.585 | -0.1495 | Yes |
| 163 | CERKL | ceramide kinase like [Source:HGNC Symbol;Acc:HGNC:21699] | 36593 | -3.642 | -0.1270 | Yes |
| 164 | NID2 | nidogen 2 [Source:HGNC Symbol;Acc:HGNC:13389] | 36793 | -3.990 | -0.1066 | Yes |
| 165 | PKP2 | plakophilin 2 [Source:HGNC Symbol;Acc:HGNC:9024] | 36828 | -4.041 | -0.0814 | Yes |
| 166 | FRMD4B | FERM domain containing 4B [Source:HGNC Symbol;Acc:HGNC:24886] | 36927 | -4.302 | -0.0563 | Yes |
| 167 | SCRN2 | secernin 2 [Source:HGNC Symbol;Acc:HGNC:30381] | 36965 | -4.404 | -0.0289 | Yes |
| 168 | CENPS | centromere protein S [Source:HGNC Symbol;Acc:HGNC:23163] | 37090 | -4.999 | 0.0000 | Yes |