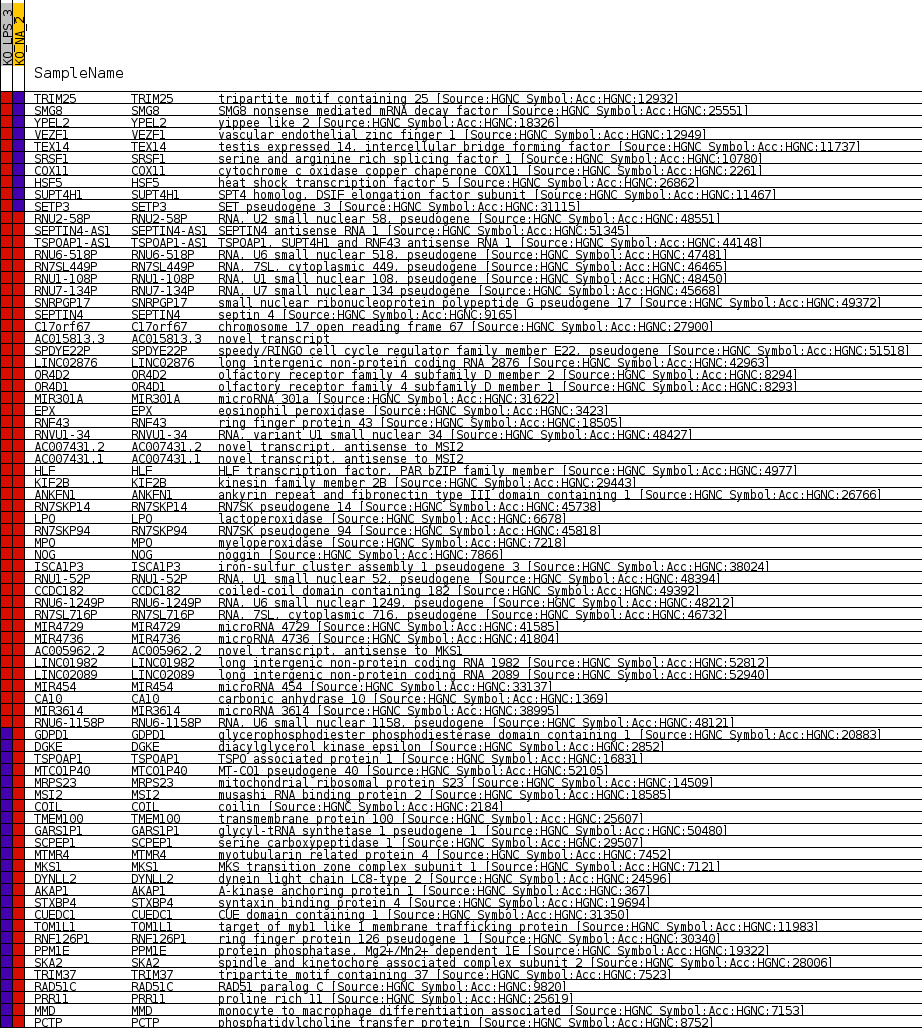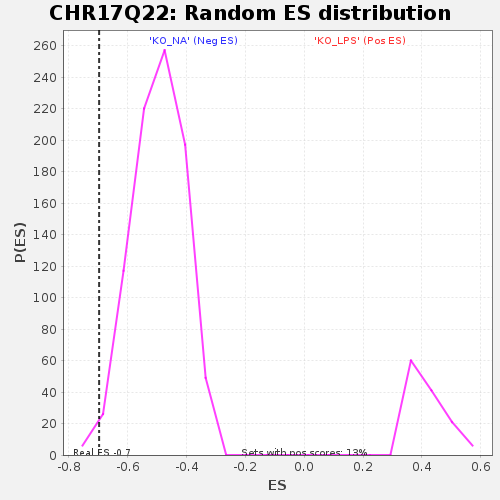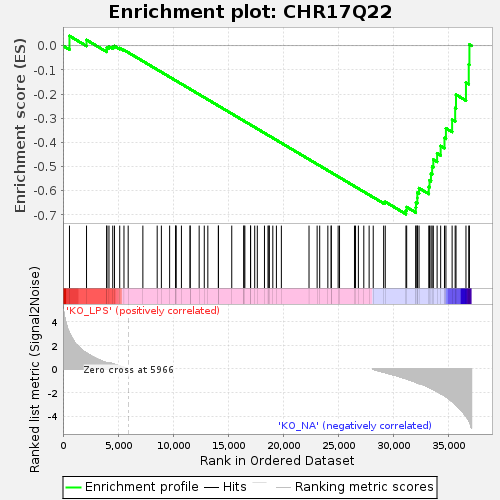
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | CHR17Q22 |
| Enrichment Score (ES) | -0.6969276 |
| Normalized Enrichment Score (NES) | -1.405803 |
| Nominal p-value | 0.010321101 |
| FDR q-value | 0.6547901 |
| FWER p-Value | 0.892 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | TRIM25 | tripartite motif containing 25 [Source:HGNC Symbol;Acc:HGNC:12932] | 588 | 3.015 | 0.0409 | No |
| 2 | SMG8 | SMG8 nonsense mediated mRNA decay factor [Source:HGNC Symbol;Acc:HGNC:25551] | 2136 | 1.334 | 0.0242 | No |
| 3 | YPEL2 | yippee like 2 [Source:HGNC Symbol;Acc:HGNC:18326] | 3964 | 0.502 | -0.0157 | No |
| 4 | VEZF1 | vascular endothelial zinc finger 1 [Source:HGNC Symbol;Acc:HGNC:12949] | 3984 | 0.496 | -0.0069 | No |
| 5 | TEX14 | testis expressed 14, intercellular bridge forming factor [Source:HGNC Symbol;Acc:HGNC:11737] | 4173 | 0.473 | -0.0030 | No |
| 6 | SRSF1 | serine and arginine rich splicing factor 1 [Source:HGNC Symbol;Acc:HGNC:10780] | 4515 | 0.437 | -0.0040 | No |
| 7 | COX11 | cytochrome c oxidase copper chaperone COX11 [Source:HGNC Symbol;Acc:HGNC:2261] | 4673 | 0.382 | -0.0011 | No |
| 8 | HSF5 | heat shock transcription factor 5 [Source:HGNC Symbol;Acc:HGNC:26862] | 5149 | 0.231 | -0.0095 | No |
| 9 | SUPT4H1 | SPT4 homolog, DSIF elongation factor subunit [Source:HGNC Symbol;Acc:HGNC:11467] | 5535 | 0.126 | -0.0176 | No |
| 10 | SETP3 | SET pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:31115] | 5917 | 0.012 | -0.0276 | No |
| 11 | RNU2-58P | RNA, U2 small nuclear 58, pseudogene [Source:HGNC Symbol;Acc:HGNC:48551] | 7250 | 0.000 | -0.0636 | No |
| 12 | SEPTIN4-AS1 | SEPTIN4 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:51345] | 8550 | 0.000 | -0.0987 | No |
| 13 | TSPOAP1-AS1 | TSPOAP1, SUPT4H1 and RNF43 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:44148] | 8931 | 0.000 | -0.1090 | No |
| 14 | RNU6-518P | RNA, U6 small nuclear 518, pseudogene [Source:HGNC Symbol;Acc:HGNC:47481] | 9680 | 0.000 | -0.1292 | No |
| 15 | RN7SL449P | RNA, 7SL, cytoplasmic 449, pseudogene [Source:HGNC Symbol;Acc:HGNC:46465] | 10228 | 0.000 | -0.1440 | No |
| 16 | RNU1-108P | RNA, U1 small nuclear 108, pseudogene [Source:HGNC Symbol;Acc:HGNC:48450] | 10238 | 0.000 | -0.1442 | No |
| 17 | RNU7-134P | RNA, U7 small nuclear 134 pseudogene [Source:HGNC Symbol;Acc:HGNC:45668] | 10261 | 0.000 | -0.1448 | No |
| 18 | SNRPGP17 | small nuclear ribonucleoprotein polypeptide G pseudogene 17 [Source:HGNC Symbol;Acc:HGNC:49372] | 10751 | 0.000 | -0.1580 | No |
| 19 | SEPTIN4 | septin 4 [Source:HGNC Symbol;Acc:HGNC:9165] | 11536 | 0.000 | -0.1792 | No |
| 20 | C17orf67 | chromosome 17 open reading frame 67 [Source:HGNC Symbol;Acc:HGNC:27900] | 11541 | 0.000 | -0.1793 | No |
| 21 | AC015813.3 | novel transcript | 12360 | 0.000 | -0.2014 | No |
| 22 | SPDYE22P | speedy/RINGO cell cycle regulator family member E22, pseudogene [Source:HGNC Symbol;Acc:HGNC:51518] | 12825 | 0.000 | -0.2140 | No |
| 23 | LINC02876 | long intergenic non-protein coding RNA 2876 [Source:HGNC Symbol;Acc:HGNC:42963] | 13148 | 0.000 | -0.2227 | No |
| 24 | OR4D2 | olfactory receptor family 4 subfamily D member 2 [Source:HGNC Symbol;Acc:HGNC:8294] | 14107 | 0.000 | -0.2485 | No |
| 25 | OR4D1 | olfactory receptor family 4 subfamily D member 1 [Source:HGNC Symbol;Acc:HGNC:8293] | 14108 | 0.000 | -0.2485 | No |
| 26 | MIR301A | microRNA 301a [Source:HGNC Symbol;Acc:HGNC:31622] | 15319 | 0.000 | -0.2812 | No |
| 27 | EPX | eosinophil peroxidase [Source:HGNC Symbol;Acc:HGNC:3423] | 16415 | 0.000 | -0.3108 | No |
| 28 | RNF43 | ring finger protein 43 [Source:HGNC Symbol;Acc:HGNC:18505] | 16421 | 0.000 | -0.3110 | No |
| 29 | RNVU1-34 | RNA, variant U1 small nuclear 34 [Source:HGNC Symbol;Acc:HGNC:48427] | 16509 | 0.000 | -0.3133 | No |
| 30 | AC007431.2 | novel transcript, antisense to MSI2 | 17026 | 0.000 | -0.3272 | No |
| 31 | AC007431.1 | novel transcript, antisense to MSI2 | 17029 | 0.000 | -0.3273 | No |
| 32 | HLF | HLF transcription factor, PAR bZIP family member [Source:HGNC Symbol;Acc:HGNC:4977] | 17403 | 0.000 | -0.3374 | No |
| 33 | KIF2B | kinesin family member 2B [Source:HGNC Symbol;Acc:HGNC:29443] | 17639 | 0.000 | -0.3437 | No |
| 34 | ANKFN1 | ankyrin repeat and fibronectin type III domain containing 1 [Source:HGNC Symbol;Acc:HGNC:26766] | 18289 | 0.000 | -0.3613 | No |
| 35 | RN7SKP14 | RN7SK pseudogene 14 [Source:HGNC Symbol;Acc:HGNC:45738] | 18610 | 0.000 | -0.3699 | No |
| 36 | LPO | lactoperoxidase [Source:HGNC Symbol;Acc:HGNC:6678] | 18693 | 0.000 | -0.3721 | No |
| 37 | RN7SKP94 | RN7SK pseudogene 94 [Source:HGNC Symbol;Acc:HGNC:45818] | 18727 | 0.000 | -0.3730 | No |
| 38 | MPO | myeloperoxidase [Source:HGNC Symbol;Acc:HGNC:7218] | 19047 | 0.000 | -0.3816 | No |
| 39 | NOG | noggin [Source:HGNC Symbol;Acc:HGNC:7866] | 19373 | 0.000 | -0.3904 | No |
| 40 | ISCA1P3 | iron-sulfur cluster assembly 1 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:38024] | 19823 | 0.000 | -0.4025 | No |
| 41 | RNU1-52P | RNA, U1 small nuclear 52, pseudogene [Source:HGNC Symbol;Acc:HGNC:48394] | 22333 | 0.000 | -0.4703 | No |
| 42 | CCDC182 | coiled-coil domain containing 182 [Source:HGNC Symbol;Acc:HGNC:49392] | 23070 | 0.000 | -0.4902 | No |
| 43 | RNU6-1249P | RNA, U6 small nuclear 1249, pseudogene [Source:HGNC Symbol;Acc:HGNC:48212] | 23303 | 0.000 | -0.4965 | No |
| 44 | RN7SL716P | RNA, 7SL, cytoplasmic 716, pseudogene [Source:HGNC Symbol;Acc:HGNC:46732] | 24042 | 0.000 | -0.5164 | No |
| 45 | MIR4729 | microRNA 4729 [Source:HGNC Symbol;Acc:HGNC:41585] | 24345 | 0.000 | -0.5246 | No |
| 46 | MIR4736 | microRNA 4736 [Source:HGNC Symbol;Acc:HGNC:41804] | 24350 | 0.000 | -0.5247 | No |
| 47 | AC005962.2 | novel transcript, antisense to MKS1 | 24977 | 0.000 | -0.5416 | No |
| 48 | LINC01982 | long intergenic non-protein coding RNA 1982 [Source:HGNC Symbol;Acc:HGNC:52812] | 25103 | 0.000 | -0.5450 | No |
| 49 | LINC02089 | long intergenic non-protein coding RNA 2089 [Source:HGNC Symbol;Acc:HGNC:52940] | 26460 | 0.000 | -0.5816 | No |
| 50 | MIR454 | microRNA 454 [Source:HGNC Symbol;Acc:HGNC:33137] | 26556 | 0.000 | -0.5842 | No |
| 51 | CA10 | carbonic anhydrase 10 [Source:HGNC Symbol;Acc:HGNC:1369] | 26815 | 0.000 | -0.5912 | No |
| 52 | MIR3614 | microRNA 3614 [Source:HGNC Symbol;Acc:HGNC:38995] | 27306 | 0.000 | -0.6044 | No |
| 53 | RNU6-1158P | RNA, U6 small nuclear 1158, pseudogene [Source:HGNC Symbol;Acc:HGNC:48121] | 27790 | 0.000 | -0.6174 | No |
| 54 | GDPD1 | glycerophosphodiester phosphodiesterase domain containing 1 [Source:HGNC Symbol;Acc:HGNC:20883] | 28162 | -0.013 | -0.6272 | No |
| 55 | DGKE | diacylglycerol kinase epsilon [Source:HGNC Symbol;Acc:HGNC:2852] | 29108 | -0.256 | -0.6479 | No |
| 56 | TSPOAP1 | TSPO associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16831] | 29254 | -0.299 | -0.6462 | No |
| 57 | MTCO1P40 | MT-CO1 pseudogene 40 [Source:HGNC Symbol;Acc:HGNC:52105] | 31132 | -0.803 | -0.6818 | Yes |
| 58 | MRPS23 | mitochondrial ribosomal protein S23 [Source:HGNC Symbol;Acc:HGNC:14509] | 31199 | -0.826 | -0.6680 | Yes |
| 59 | MSI2 | musashi RNA binding protein 2 [Source:HGNC Symbol;Acc:HGNC:18585] | 32025 | -1.103 | -0.6696 | Yes |
| 60 | COIL | coilin [Source:HGNC Symbol;Acc:HGNC:2184] | 32046 | -1.111 | -0.6492 | Yes |
| 61 | TMEM100 | transmembrane protein 100 [Source:HGNC Symbol;Acc:HGNC:25607] | 32163 | -1.163 | -0.6304 | Yes |
| 62 | GARS1P1 | glycyl-tRNA synthetase 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:50480] | 32167 | -1.165 | -0.6086 | Yes |
| 63 | SCPEP1 | serine carboxypeptidase 1 [Source:HGNC Symbol;Acc:HGNC:29507] | 32321 | -1.230 | -0.5895 | Yes |
| 64 | MTMR4 | myotubularin related protein 4 [Source:HGNC Symbol;Acc:HGNC:7452] | 33207 | -1.538 | -0.5845 | Yes |
| 65 | MKS1 | MKS transition zone complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:7121] | 33275 | -1.564 | -0.5568 | Yes |
| 66 | DYNLL2 | dynein light chain LC8-type 2 [Source:HGNC Symbol;Acc:HGNC:24596] | 33405 | -1.625 | -0.5297 | Yes |
| 67 | AKAP1 | A-kinase anchoring protein 1 [Source:HGNC Symbol;Acc:HGNC:367] | 33509 | -1.668 | -0.5011 | Yes |
| 68 | STXBP4 | syntaxin binding protein 4 [Source:HGNC Symbol;Acc:HGNC:19694] | 33617 | -1.729 | -0.4714 | Yes |
| 69 | CUEDC1 | CUE domain containing 1 [Source:HGNC Symbol;Acc:HGNC:31350] | 33962 | -1.891 | -0.4451 | Yes |
| 70 | TOM1L1 | target of myb1 like 1 membrane trafficking protein [Source:HGNC Symbol;Acc:HGNC:11983] | 34284 | -2.068 | -0.4148 | Yes |
| 71 | RNF126P1 | ring finger protein 126 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:30340] | 34631 | -2.251 | -0.3818 | Yes |
| 72 | PPM1E | protein phosphatase, Mg2+/Mn2+ dependent 1E [Source:HGNC Symbol;Acc:HGNC:19322] | 34762 | -2.321 | -0.3416 | Yes |
| 73 | SKA2 | spindle and kinetochore associated complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:28006] | 35324 | -2.700 | -0.3059 | Yes |
| 74 | TRIM37 | tripartite motif containing 37 [Source:HGNC Symbol;Acc:HGNC:7523] | 35600 | -2.947 | -0.2578 | Yes |
| 75 | RAD51C | RAD51 paralog C [Source:HGNC Symbol;Acc:HGNC:9820] | 35677 | -3.050 | -0.2025 | Yes |
| 76 | PRR11 | proline rich 11 [Source:HGNC Symbol;Acc:HGNC:25619] | 36581 | -3.976 | -0.1520 | Yes |
| 77 | MMD | monocyte to macrophage differentiation associated [Source:HGNC Symbol;Acc:HGNC:7153] | 36835 | -4.346 | -0.0770 | Yes |
| 78 | PCTP | phosphatidylcholine transfer protein [Source:HGNC Symbol;Acc:HGNC:8752] | 36894 | -4.451 | 0.0053 | Yes |