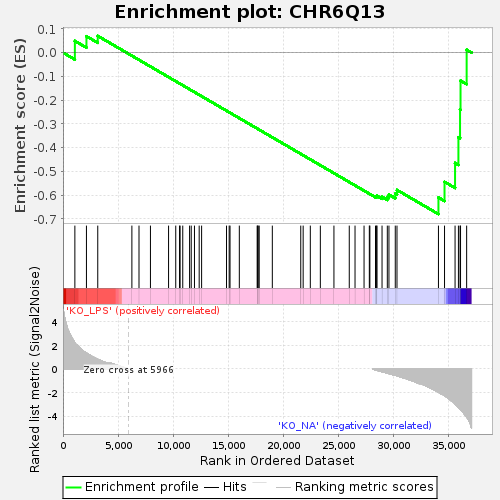
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
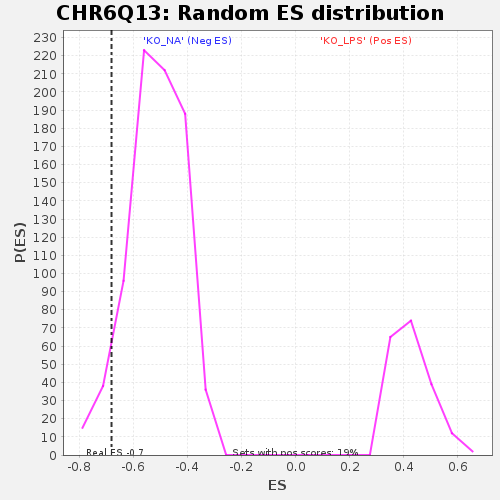
| GeneSet | CHR6Q13 |
| Enrichment Score (ES) | -0.6800154 |
| Normalized Enrichment Score (NES) | -1.3183854 |
| Nominal p-value | 0.063118815 |
| FDR q-value | 0.6340036 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CGAS | cyclic GMP-AMP synthase [Source:HGNC Symbol;Acc:HGNC:21367] | 1077 | 2.189 | 0.0493 | No |
| 2 | TXNP7 | thioredoxin pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:49487] | 2128 | 1.342 | 0.0689 | No |
| 3 | LMBRD1 | LMBR1 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:23038] | 3155 | 0.814 | 0.0704 | No |
| 4 | B3GAT2 | beta-1,3-glucuronyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:922] | 6253 | 0.000 | -0.0132 | No |
| 5 | LINC01626 | long intergenic non-protein coding RNA 1626 [Source:HGNC Symbol;Acc:HGNC:52257] | 6899 | 0.000 | -0.0306 | No |
| 6 | DPPA5 | developmental pluripotency associated 5 [Source:HGNC Symbol;Acc:HGNC:19201] | 7938 | 0.000 | -0.0587 | No |
| 7 | KCNQ5 | potassium voltage-gated channel subfamily Q member 5 [Source:HGNC Symbol;Acc:HGNC:6299] | 9583 | 0.000 | -0.1030 | No |
| 8 | KRT19P1 | keratin 19 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:33422] | 10235 | 0.000 | -0.1206 | No |
| 9 | NDUFAB1P1 | NADH:ubiquinone oxidoreductase subunit AB1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:52265] | 10580 | 0.000 | -0.1299 | No |
| 10 | GAPDHP42 | glyceraldehyde 3 phosphate dehydrogenase pseudogene 42 [Source:HGNC Symbol;Acc:HGNC:37799] | 10630 | 0.000 | -0.1312 | No |
| 11 | OOEP-AS1 | OOEP antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:39192] | 10871 | 0.000 | -0.1377 | No |
| 12 | RBPMS2P1 | RBPMS2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:50509] | 11492 | 0.000 | -0.1544 | No |
| 13 | RNU4-66P | RNA, U4 small nuclear 66, pseudogene [Source:HGNC Symbol;Acc:HGNC:47002] | 11641 | 0.000 | -0.1584 | No |
| 14 | KHDC1 | KH domain containing 1 [Source:HGNC Symbol;Acc:HGNC:21366] | 11934 | 0.000 | -0.1663 | No |
| 15 | KHDC1P1 | KH domain containing 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:43757] | 12362 | 0.000 | -0.1779 | No |
| 16 | SLC25A6P6 | solute carrier family 25 member 6 pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:43855] | 12583 | 0.000 | -0.1838 | No |
| 17 | RNU7-48P | RNA, U7 small nuclear 48 pseudogene [Source:HGNC Symbol;Acc:HGNC:34144] | 14844 | 0.000 | -0.2448 | No |
| 18 | MIR30A | microRNA 30a [Source:HGNC Symbol;Acc:HGNC:31624] | 15084 | 0.000 | -0.2513 | No |
| 19 | MIR30C2 | microRNA 30c-2 [Source:HGNC Symbol;Acc:HGNC:31627] | 15162 | 0.000 | -0.2533 | No |
| 20 | RNU6-411P | RNA, U6 small nuclear 411, pseudogene [Source:HGNC Symbol;Acc:HGNC:47374] | 16006 | 0.000 | -0.2761 | No |
| 21 | OOEP | oocyte expressed protein [Source:HGNC Symbol;Acc:HGNC:21382] | 17636 | 0.000 | -0.3201 | No |
| 22 | COL19A1 | collagen type XIX alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:2196] | 17644 | 0.000 | -0.3203 | No |
| 23 | BECN1P2 | beclin 1 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:51330] | 17724 | 0.000 | -0.3224 | No |
| 24 | SDCBP2P1 | syndecan binding protein 2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:44685] | 17810 | 0.000 | -0.3247 | No |
| 25 | COL9A1 | collagen type IX alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:2217] | 19002 | 0.000 | -0.3568 | No |
| 26 | LINC00472 | long intergenic non-protein coding RNA 472 [Source:HGNC Symbol;Acc:HGNC:21380] | 21583 | 0.000 | -0.4265 | No |
| 27 | AL590428.1 | novel transcript | 21802 | 0.000 | -0.4324 | No |
| 28 | RIMS1 | regulating synaptic membrane exocytosis 1 [Source:HGNC Symbol;Acc:HGNC:17282] | 22451 | 0.000 | -0.4499 | No |
| 29 | MIR4282 | microRNA 4282 [Source:HGNC Symbol;Acc:HGNC:38189] | 23361 | 0.000 | -0.4744 | No |
| 30 | AL357507.1 | novel transcript | 24595 | 0.000 | -0.5077 | No |
| 31 | KCNQ5-AS1 | KCNQ5 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:40323] | 25987 | 0.000 | -0.5453 | No |
| 32 | RNU6-975P | RNA, U6 small nuclear 975, pseudogene [Source:HGNC Symbol;Acc:HGNC:47938] | 26510 | 0.000 | -0.5594 | No |
| 33 | RPSAP41 | ribosomal protein SA pseudogene 41 [Source:HGNC Symbol;Acc:HGNC:36663] | 27329 | 0.000 | -0.5814 | No |
| 34 | KHDC1L | KH domain containing 1 like [Source:HGNC Symbol;Acc:HGNC:37274] | 27808 | 0.000 | -0.5943 | No |
| 35 | KHDC3L | KH domain containing 3 like, subcortical maternal complex member [Source:HGNC Symbol;Acc:HGNC:33699] | 27857 | 0.000 | -0.5956 | No |
| 36 | NPM1P37 | nucleophosmin 1 pseudogene 37 [Source:HGNC Symbol;Acc:HGNC:45216] | 28369 | -0.073 | -0.6068 | No |
| 37 | EIF3EP1 | eukaryotic translation initiation factor 3 subunit E pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:6102] | 28473 | -0.095 | -0.6062 | No |
| 38 | LYPLA1P3 | LYPLA1 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:44007] | 28500 | -0.105 | -0.6032 | No |
| 39 | MTO1 | mitochondrial tRNA translation optimization 1 [Source:HGNC Symbol;Acc:HGNC:19261] | 28962 | -0.226 | -0.6075 | No |
| 40 | EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 [Source:HGNC Symbol;Acc:HGNC:3189] | 29441 | -0.352 | -0.6078 | No |
| 41 | SMAP1 | small ArfGAP 1 [Source:HGNC Symbol;Acc:HGNC:19651] | 29589 | -0.384 | -0.5981 | No |
| 42 | SDHAF4 | succinate dehydrogenase complex assembly factor 4 [Source:HGNC Symbol;Acc:HGNC:20957] | 30163 | -0.537 | -0.5943 | No |
| 43 | KNOP1P4 | lysine rich nucleolar protein 1 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:48922] | 30319 | -0.561 | -0.5784 | No |
| 44 | FAM135A | family with sequence similarity 135 member A [Source:HGNC Symbol;Acc:HGNC:21084] | 34083 | -1.955 | -0.6100 | Yes |
| 45 | DDX43 | DEAD-box helicase 43 [Source:HGNC Symbol;Acc:HGNC:18677] | 34635 | -2.254 | -0.5442 | Yes |
| 46 | PGAM1P10 | phosphoglycerate mutase 1 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:42457] | 35591 | -2.942 | -0.4647 | Yes |
| 47 | PAICSP3 | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:38096] | 35896 | -3.254 | -0.3564 | Yes |
| 48 | OGFRL1 | opioid growth factor receptor like 1 [Source:HGNC Symbol;Acc:HGNC:21378] | 36044 | -3.376 | -0.2395 | Yes |
| 49 | SLC17A5 | solute carrier family 17 member 5 [Source:HGNC Symbol;Acc:HGNC:10933] | 36088 | -3.428 | -0.1180 | Yes |
| 50 | CD109 | CD109 molecule [Source:HGNC Symbol;Acc:HGNC:21685] | 36643 | -4.051 | 0.0121 | Yes |