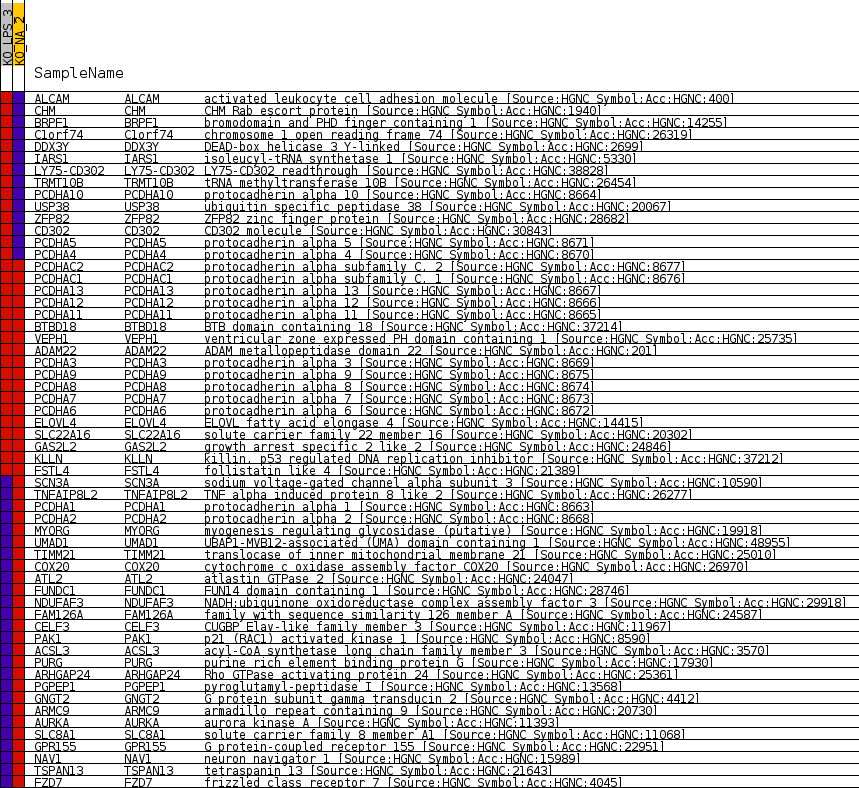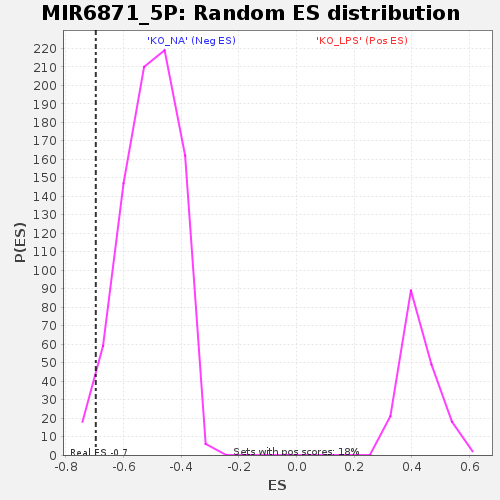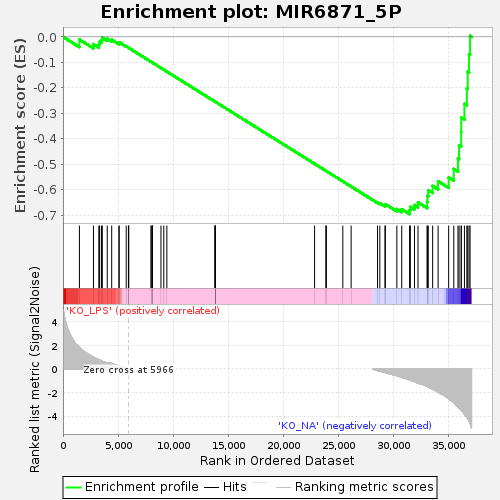
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | MIR6871_5P |
| Enrichment Score (ES) | -0.6962647 |
| Normalized Enrichment Score (NES) | -1.3712131 |
| Nominal p-value | 0.028014617 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ALCAM | activated leukocyte cell adhesion molecule [Source:HGNC Symbol;Acc:HGNC:400] | 1490 | 1.822 | -0.0105 | No |
| 2 | CHM | CHM Rab escort protein [Source:HGNC Symbol;Acc:HGNC:1940] | 2765 | 0.996 | -0.0287 | No |
| 3 | BRPF1 | bromodomain and PHD finger containing 1 [Source:HGNC Symbol;Acc:HGNC:14255] | 3253 | 0.777 | -0.0292 | No |
| 4 | C1orf74 | chromosome 1 open reading frame 74 [Source:HGNC Symbol;Acc:HGNC:26319] | 3324 | 0.752 | -0.0188 | No |
| 5 | DDX3Y | DEAD-box helicase 3 Y-linked [Source:HGNC Symbol;Acc:HGNC:2699] | 3492 | 0.685 | -0.0121 | No |
| 6 | IARS1 | isoleucyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:5330] | 3567 | 0.655 | -0.0035 | No |
| 7 | LY75-CD302 | LY75-CD302 readthrough [Source:HGNC Symbol;Acc:HGNC:38828] | 4018 | 0.481 | -0.0078 | No |
| 8 | TRMT10B | tRNA methyltransferase 10B [Source:HGNC Symbol;Acc:HGNC:26454] | 4421 | 0.465 | -0.0110 | No |
| 9 | PCDHA10 | protocadherin alpha 10 [Source:HGNC Symbol;Acc:HGNC:8664] | 5081 | 0.249 | -0.0248 | No |
| 10 | USP38 | ubiquitin specific peptidase 38 [Source:HGNC Symbol;Acc:HGNC:20067] | 5095 | 0.246 | -0.0211 | No |
| 11 | ZFP82 | ZFP82 zinc finger protein [Source:HGNC Symbol;Acc:HGNC:28682] | 5739 | 0.068 | -0.0374 | No |
| 12 | CD302 | CD302 molecule [Source:HGNC Symbol;Acc:HGNC:30843] | 5941 | 0.004 | -0.0427 | No |
| 13 | PCDHA5 | protocadherin alpha 5 [Source:HGNC Symbol;Acc:HGNC:8671] | 5957 | 0.000 | -0.0431 | No |
| 14 | PCDHA4 | protocadherin alpha 4 [Source:HGNC Symbol;Acc:HGNC:8670] | 5961 | 0.000 | -0.0432 | No |
| 15 | PCDHAC2 | protocadherin alpha subfamily C, 2 [Source:HGNC Symbol;Acc:HGNC:8677] | 8000 | 0.000 | -0.0983 | No |
| 16 | PCDHAC1 | protocadherin alpha subfamily C, 1 [Source:HGNC Symbol;Acc:HGNC:8676] | 8001 | 0.000 | -0.0983 | No |
| 17 | PCDHA13 | protocadherin alpha 13 [Source:HGNC Symbol;Acc:HGNC:8667] | 8116 | 0.000 | -0.1013 | No |
| 18 | PCDHA12 | protocadherin alpha 12 [Source:HGNC Symbol;Acc:HGNC:8666] | 8118 | 0.000 | -0.1014 | No |
| 19 | PCDHA11 | protocadherin alpha 11 [Source:HGNC Symbol;Acc:HGNC:8665] | 8120 | 0.000 | -0.1014 | No |
| 20 | BTBD18 | BTB domain containing 18 [Source:HGNC Symbol;Acc:HGNC:37214] | 8893 | 0.000 | -0.1222 | No |
| 21 | VEPH1 | ventricular zone expressed PH domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25735] | 9149 | 0.000 | -0.1291 | No |
| 22 | ADAM22 | ADAM metallopeptidase domain 22 [Source:HGNC Symbol;Acc:HGNC:201] | 9428 | 0.000 | -0.1366 | No |
| 23 | PCDHA3 | protocadherin alpha 3 [Source:HGNC Symbol;Acc:HGNC:8669] | 13817 | 0.000 | -0.2551 | No |
| 24 | PCDHA9 | protocadherin alpha 9 [Source:HGNC Symbol;Acc:HGNC:8675] | 13818 | 0.000 | -0.2551 | No |
| 25 | PCDHA8 | protocadherin alpha 8 [Source:HGNC Symbol;Acc:HGNC:8674] | 13819 | 0.000 | -0.2551 | No |
| 26 | PCDHA7 | protocadherin alpha 7 [Source:HGNC Symbol;Acc:HGNC:8673] | 13820 | 0.000 | -0.2551 | No |
| 27 | PCDHA6 | protocadherin alpha 6 [Source:HGNC Symbol;Acc:HGNC:8672] | 13821 | 0.000 | -0.2551 | No |
| 28 | ELOVL4 | ELOVL fatty acid elongase 4 [Source:HGNC Symbol;Acc:HGNC:14415] | 22838 | 0.000 | -0.4986 | No |
| 29 | SLC22A16 | solute carrier family 22 member 16 [Source:HGNC Symbol;Acc:HGNC:20302] | 23857 | 0.000 | -0.5261 | No |
| 30 | GAS2L2 | growth arrest specific 2 like 2 [Source:HGNC Symbol;Acc:HGNC:24846] | 23914 | 0.000 | -0.5276 | No |
| 31 | KLLN | killin, p53 regulated DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:37212] | 25403 | 0.000 | -0.5678 | No |
| 32 | FSTL4 | follistatin like 4 [Source:HGNC Symbol;Acc:HGNC:21389] | 26156 | 0.000 | -0.5881 | No |
| 33 | SCN3A | sodium voltage-gated channel alpha subunit 3 [Source:HGNC Symbol;Acc:HGNC:10590] | 28553 | -0.119 | -0.6508 | No |
| 34 | TNFAIP8L2 | TNF alpha induced protein 8 like 2 [Source:HGNC Symbol;Acc:HGNC:26277] | 28761 | -0.172 | -0.6536 | No |
| 35 | PCDHA1 | protocadherin alpha 1 [Source:HGNC Symbol;Acc:HGNC:8663] | 29229 | -0.292 | -0.6615 | No |
| 36 | PCDHA2 | protocadherin alpha 2 [Source:HGNC Symbol;Acc:HGNC:8668] | 29264 | -0.302 | -0.6575 | No |
| 37 | MYORG | myogenesis regulating glycosidase (putative) [Source:HGNC Symbol;Acc:HGNC:19918] | 30308 | -0.558 | -0.6765 | No |
| 38 | UMAD1 | UBAP1-MVB12-associated (UMA) domain containing 1 [Source:HGNC Symbol;Acc:HGNC:48955] | 30750 | -0.696 | -0.6771 | No |
| 39 | TIMM21 | translocase of inner mitochondrial membrane 21 [Source:HGNC Symbol;Acc:HGNC:25010] | 31461 | -0.914 | -0.6814 | Yes |
| 40 | COX20 | cytochrome c oxidase assembly factor COX20 [Source:HGNC Symbol;Acc:HGNC:26970] | 31532 | -0.929 | -0.6681 | Yes |
| 41 | ATL2 | atlastin GTPase 2 [Source:HGNC Symbol;Acc:HGNC:24047] | 31905 | -1.060 | -0.6609 | Yes |
| 42 | FUNDC1 | FUN14 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:28746] | 32227 | -1.189 | -0.6502 | Yes |
| 43 | NDUFAF3 | NADH:ubiquinone oxidoreductase complex assembly factor 3 [Source:HGNC Symbol;Acc:HGNC:29918] | 33055 | -1.476 | -0.6484 | Yes |
| 44 | FAM126A | family with sequence similarity 126 member A [Source:HGNC Symbol;Acc:HGNC:24587] | 33076 | -1.484 | -0.6248 | Yes |
| 45 | CELF3 | CUGBP Elav-like family member 3 [Source:HGNC Symbol;Acc:HGNC:11967] | 33159 | -1.513 | -0.6024 | Yes |
| 46 | PAK1 | p21 (RAC1) activated kinase 1 [Source:HGNC Symbol;Acc:HGNC:8590] | 33557 | -1.697 | -0.5854 | Yes |
| 47 | ACSL3 | acyl-CoA synthetase long chain family member 3 [Source:HGNC Symbol;Acc:HGNC:3570] | 34050 | -1.939 | -0.5671 | Yes |
| 48 | PURG | purine rich element binding protein G [Source:HGNC Symbol;Acc:HGNC:17930] | 35010 | -2.492 | -0.5524 | Yes |
| 49 | ARHGAP24 | Rho GTPase activating protein 24 [Source:HGNC Symbol;Acc:HGNC:25361] | 35475 | -2.815 | -0.5190 | Yes |
| 50 | PGPEP1 | pyroglutamyl-peptidase I [Source:HGNC Symbol;Acc:HGNC:13568] | 35855 | -3.192 | -0.4772 | Yes |
| 51 | GNGT2 | G protein subunit gamma transducin 2 [Source:HGNC Symbol;Acc:HGNC:4412] | 35955 | -3.321 | -0.4257 | Yes |
| 52 | ARMC9 | armadillo repeat containing 9 [Source:HGNC Symbol;Acc:HGNC:20730] | 36149 | -3.485 | -0.3741 | Yes |
| 53 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 36154 | -3.490 | -0.3174 | Yes |
| 54 | SLC8A1 | solute carrier family 8 member A1 [Source:HGNC Symbol;Acc:HGNC:11068] | 36441 | -3.802 | -0.2631 | Yes |
| 55 | GPR155 | G protein-coupled receptor 155 [Source:HGNC Symbol;Acc:HGNC:22951] | 36661 | -4.067 | -0.2027 | Yes |
| 56 | NAV1 | neuron navigator 1 [Source:HGNC Symbol;Acc:HGNC:15989] | 36739 | -4.177 | -0.1367 | Yes |
| 57 | TSPAN13 | tetraspanin 13 [Source:HGNC Symbol;Acc:HGNC:21643] | 36862 | -4.397 | -0.0683 | Yes |
| 58 | FZD7 | frizzled class receptor 7 [Source:HGNC Symbol;Acc:HGNC:4045] | 36953 | -4.567 | 0.0037 | Yes |