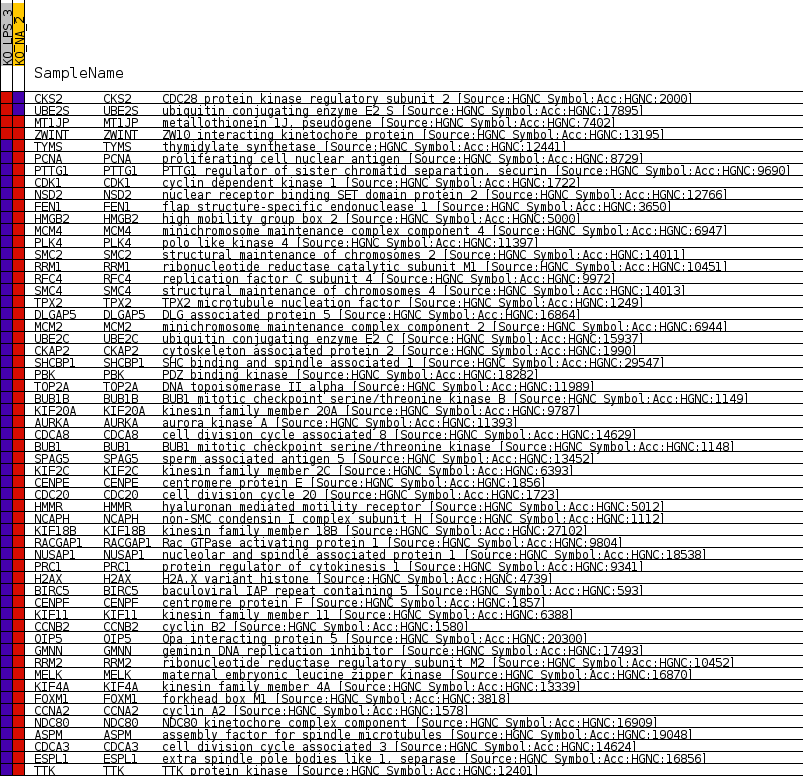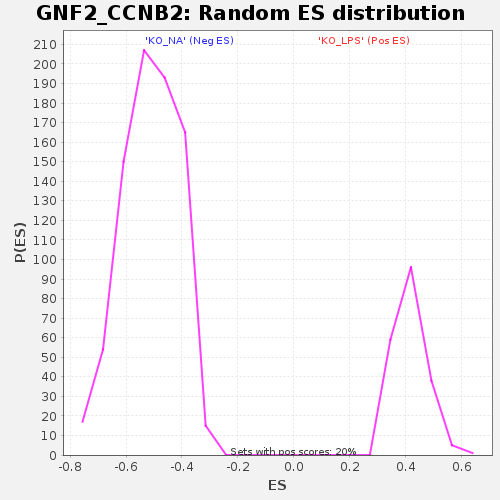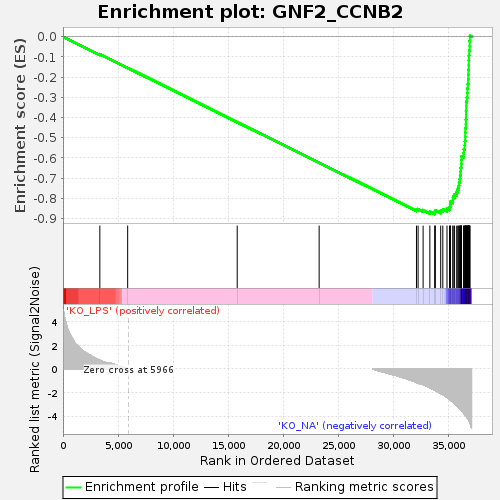
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_CCNB2 |
| Enrichment Score (ES) | -0.8776771 |
| Normalized Enrichment Score (NES) | -1.7207068 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 3345 | 0.744 | -0.0862 | No |
| 2 | UBE2S | ubiquitin conjugating enzyme E2 S [Source:HGNC Symbol;Acc:HGNC:17895] | 5870 | 0.021 | -0.1542 | No |
| 3 | MT1JP | metallothionein 1J, pseudogene [Source:HGNC Symbol;Acc:HGNC:7402] | 15814 | 0.000 | -0.4227 | No |
| 4 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 23253 | 0.000 | -0.6236 | No |
| 5 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32088 | -1.131 | -0.8558 | No |
| 6 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 32234 | -1.194 | -0.8531 | No |
| 7 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 32691 | -1.316 | -0.8581 | No |
| 8 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 33303 | -1.578 | -0.8659 | No |
| 9 | NSD2 | nuclear receptor binding SET domain protein 2 [Source:HGNC Symbol;Acc:HGNC:12766] | 33740 | -1.773 | -0.8679 | Yes |
| 10 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 33804 | -1.813 | -0.8595 | Yes |
| 11 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 34287 | -2.069 | -0.8611 | Yes |
| 12 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34480 | -2.139 | -0.8544 | Yes |
| 13 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34858 | -2.395 | -0.8513 | Yes |
| 14 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35087 | -2.553 | -0.8433 | Yes |
| 15 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35160 | -2.606 | -0.8308 | Yes |
| 16 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 35172 | -2.617 | -0.8166 | Yes |
| 17 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 35393 | -2.750 | -0.8073 | Yes |
| 18 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 35409 | -2.763 | -0.7924 | Yes |
| 19 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 35546 | -2.882 | -0.7801 | Yes |
| 20 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 35740 | -3.078 | -0.7683 | Yes |
| 21 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 35837 | -3.176 | -0.7533 | Yes |
| 22 | CKAP2 | cytoskeleton associated protein 2 [Source:HGNC Symbol;Acc:HGNC:1990] | 35926 | -3.289 | -0.7374 | Yes |
| 23 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 35958 | -3.323 | -0.7198 | Yes |
| 24 | PBK | PDZ binding kinase [Source:HGNC Symbol;Acc:HGNC:18282] | 36027 | -3.367 | -0.7030 | Yes |
| 25 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 36054 | -3.383 | -0.6850 | Yes |
| 26 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 36096 | -3.434 | -0.6671 | Yes |
| 27 | KIF20A | kinesin family member 20A [Source:HGNC Symbol;Acc:HGNC:9787] | 36110 | -3.445 | -0.6483 | Yes |
| 28 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 36154 | -3.490 | -0.6301 | Yes |
| 29 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 36163 | -3.495 | -0.6110 | Yes |
| 30 | BUB1 | BUB1 mitotic checkpoint serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1148] | 36172 | -3.498 | -0.5918 | Yes |
| 31 | SPAG5 | sperm associated antigen 5 [Source:HGNC Symbol;Acc:HGNC:13452] | 36366 | -3.704 | -0.5765 | Yes |
| 32 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 36388 | -3.732 | -0.5564 | Yes |
| 33 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 36469 | -3.830 | -0.5373 | Yes |
| 34 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 36488 | -3.857 | -0.5165 | Yes |
| 35 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 36512 | -3.898 | -0.4955 | Yes |
| 36 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 36519 | -3.907 | -0.4740 | Yes |
| 37 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 36526 | -3.918 | -0.4525 | Yes |
| 38 | RACGAP1 | Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] | 36572 | -3.970 | -0.4317 | Yes |
| 39 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36576 | -3.975 | -0.4097 | Yes |
| 40 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 36590 | -3.998 | -0.3879 | Yes |
| 41 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36591 | -3.999 | -0.3658 | Yes |
| 42 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 36605 | -4.013 | -0.3439 | Yes |
| 43 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36606 | -4.015 | -0.3216 | Yes |
| 44 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 36668 | -4.079 | -0.3007 | Yes |
| 45 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36709 | -4.137 | -0.2788 | Yes |
| 46 | OIP5 | Opa interacting protein 5 [Source:HGNC Symbol;Acc:HGNC:20300] | 36718 | -4.146 | -0.2561 | Yes |
| 47 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 36753 | -4.198 | -0.2337 | Yes |
| 48 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36786 | -4.241 | -0.2111 | Yes |
| 49 | MELK | maternal embryonic leucine zipper kinase [Source:HGNC Symbol;Acc:HGNC:16870] | 36795 | -4.256 | -0.1877 | Yes |
| 50 | KIF4A | kinesin family member 4A [Source:HGNC Symbol;Acc:HGNC:13339] | 36803 | -4.278 | -0.1642 | Yes |
| 51 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36833 | -4.338 | -0.1410 | Yes |
| 52 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36838 | -4.351 | -0.1170 | Yes |
| 53 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36851 | -4.372 | -0.0931 | Yes |
| 54 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36876 | -4.426 | -0.0692 | Yes |
| 55 | CDCA3 | cell division cycle associated 3 [Source:HGNC Symbol;Acc:HGNC:14624] | 36915 | -4.481 | -0.0454 | Yes |
| 56 | ESPL1 | extra spindle pole bodies like 1, separase [Source:HGNC Symbol;Acc:HGNC:16856] | 36917 | -4.488 | -0.0206 | Yes |
| 57 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 36941 | -4.548 | 0.0040 | Yes |