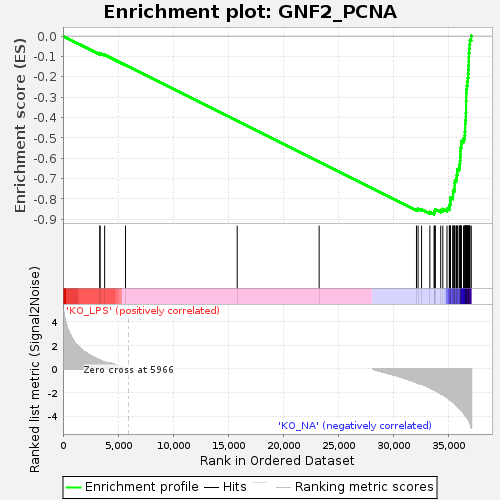
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_PCNA |
| Enrichment Score (ES) | -0.8743962 |
| Normalized Enrichment Score (NES) | -1.738401 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 3345 | 0.744 | -0.0867 | No |
| 2 | HNRNPAB | heterogeneous nuclear ribonucleoprotein A/B [Source:HGNC Symbol;Acc:HGNC:5034] | 3364 | 0.733 | -0.0837 | No |
| 3 | PA2G4 | proliferation-associated 2G4 [Source:HGNC Symbol;Acc:HGNC:8550] | 3780 | 0.570 | -0.0921 | No |
| 4 | SLBP | stem-loop binding protein [Source:HGNC Symbol;Acc:HGNC:10904] | 5676 | 0.088 | -0.1429 | No |
| 5 | MT1JP | metallothionein 1J, pseudogene [Source:HGNC Symbol;Acc:HGNC:7402] | 15814 | 0.000 | -0.4167 | No |
| 6 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 23253 | 0.000 | -0.6176 | No |
| 7 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32088 | -1.131 | -0.8507 | No |
| 8 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 32234 | -1.194 | -0.8489 | No |
| 9 | VRK1 | VRK serine/threonine kinase 1 [Source:HGNC Symbol;Acc:HGNC:12718] | 32551 | -1.246 | -0.8514 | No |
| 10 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 33303 | -1.578 | -0.8641 | No |
| 11 | PAXIP1 | PAX interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:8624] | 33687 | -1.745 | -0.8660 | Yes |
| 12 | NSD2 | nuclear receptor binding SET domain protein 2 [Source:HGNC Symbol;Acc:HGNC:12766] | 33740 | -1.773 | -0.8588 | Yes |
| 13 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 33804 | -1.813 | -0.8517 | Yes |
| 14 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 34287 | -2.069 | -0.8547 | Yes |
| 15 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34480 | -2.139 | -0.8496 | Yes |
| 16 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34858 | -2.395 | -0.8482 | Yes |
| 17 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35069 | -2.538 | -0.8416 | Yes |
| 18 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35087 | -2.553 | -0.8297 | Yes |
| 19 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 35150 | -2.598 | -0.8188 | Yes |
| 20 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35160 | -2.606 | -0.8064 | Yes |
| 21 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 35172 | -2.617 | -0.7941 | Yes |
| 22 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 35393 | -2.750 | -0.7867 | Yes |
| 23 | MCM5 | minichromosome maintenance complex component 5 [Source:HGNC Symbol;Acc:HGNC:6948] | 35403 | -2.757 | -0.7736 | Yes |
| 24 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 35409 | -2.763 | -0.7604 | Yes |
| 25 | POLE2 | DNA polymerase epsilon 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:9178] | 35534 | -2.879 | -0.7498 | Yes |
| 26 | DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 [Source:HGNC Symbol;Acc:HGNC:19123] | 35545 | -2.882 | -0.7362 | Yes |
| 27 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 35546 | -2.882 | -0.7222 | Yes |
| 28 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 35585 | -2.932 | -0.7091 | Yes |
| 29 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 35722 | -3.059 | -0.6980 | Yes |
| 30 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 35740 | -3.078 | -0.6835 | Yes |
| 31 | MCM6 | minichromosome maintenance complex component 6 [Source:HGNC Symbol;Acc:HGNC:6949] | 35783 | -3.122 | -0.6696 | Yes |
| 32 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35788 | -3.128 | -0.6545 | Yes |
| 33 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 35958 | -3.323 | -0.6430 | Yes |
| 34 | RAD54L | RAD54 like [Source:HGNC Symbol;Acc:HGNC:9826] | 36017 | -3.355 | -0.6284 | Yes |
| 35 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36039 | -3.372 | -0.6127 | Yes |
| 36 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 36054 | -3.383 | -0.5967 | Yes |
| 37 | DTL | denticleless E3 ubiquitin protein ligase homolog [Source:HGNC Symbol;Acc:HGNC:30288] | 36080 | -3.411 | -0.5808 | Yes |
| 38 | TMPO | thymopoietin [Source:HGNC Symbol;Acc:HGNC:11875] | 36086 | -3.421 | -0.5644 | Yes |
| 39 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 36096 | -3.434 | -0.5481 | Yes |
| 40 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 36154 | -3.490 | -0.5327 | Yes |
| 41 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 36163 | -3.495 | -0.5161 | Yes |
| 42 | RAD51AP1 | RAD51 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16956] | 36360 | -3.688 | -0.5035 | Yes |
| 43 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 36469 | -3.830 | -0.4879 | Yes |
| 44 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 36488 | -3.857 | -0.4697 | Yes |
| 45 | KIF15 | kinesin family member 15 [Source:HGNC Symbol;Acc:HGNC:17273] | 36511 | -3.893 | -0.4515 | Yes |
| 46 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 36512 | -3.898 | -0.4327 | Yes |
| 47 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 36526 | -3.918 | -0.4141 | Yes |
| 48 | RACGAP1 | Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] | 36572 | -3.970 | -0.3961 | Yes |
| 49 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36576 | -3.975 | -0.3770 | Yes |
| 50 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36588 | -3.994 | -0.3579 | Yes |
| 51 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 36590 | -3.998 | -0.3386 | Yes |
| 52 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36591 | -3.999 | -0.3193 | Yes |
| 53 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 36605 | -4.013 | -0.3002 | Yes |
| 54 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36606 | -4.015 | -0.2808 | Yes |
| 55 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 36619 | -4.031 | -0.2617 | Yes |
| 56 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 36668 | -4.079 | -0.2432 | Yes |
| 57 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36709 | -4.137 | -0.2243 | Yes |
| 58 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 36753 | -4.198 | -0.2052 | Yes |
| 59 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36786 | -4.241 | -0.1855 | Yes |
| 60 | MELK | maternal embryonic leucine zipper kinase [Source:HGNC Symbol;Acc:HGNC:16870] | 36795 | -4.256 | -0.1652 | Yes |
| 61 | MSH2 | mutS homolog 2 [Source:HGNC Symbol;Acc:HGNC:7325] | 36821 | -4.303 | -0.1450 | Yes |
| 62 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36833 | -4.338 | -0.1243 | Yes |
| 63 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36838 | -4.351 | -0.1034 | Yes |
| 64 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36851 | -4.372 | -0.0826 | Yes |
| 65 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36876 | -4.426 | -0.0618 | Yes |
| 66 | ESPL1 | extra spindle pole bodies like 1, separase [Source:HGNC Symbol;Acc:HGNC:16856] | 36917 | -4.488 | -0.0412 | Yes |
| 67 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 36941 | -4.548 | -0.0198 | Yes |
| 68 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 37062 | -4.928 | 0.0008 | Yes |