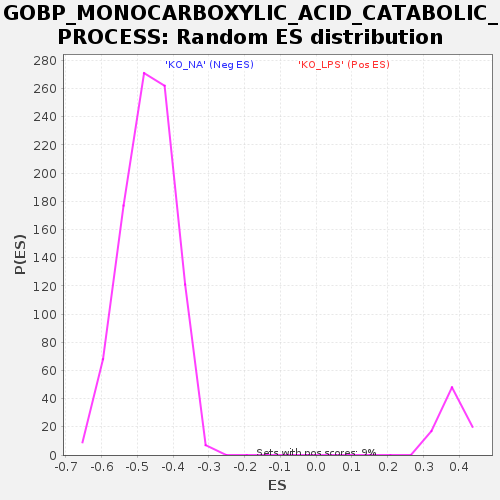
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GOBP_MONOCARBOXYLIC_ACID_CATABOLIC_PROCESS |
| Enrichment Score (ES) | -0.74094474 |
| Normalized Enrichment Score (NES) | -1.5777742 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13018921 |
| FWER p-Value | 0.858 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | HAO1 | hydroxyacid oxidase 1 [Source:HGNC Symbol;Acc:HGNC:4809] | 387 | 3.527 | 0.0078 | No |
| 2 | CYP26C1 | cytochrome P450 family 26 subfamily C member 1 [Source:HGNC Symbol;Acc:HGNC:20577] | 452 | 3.287 | 0.0231 | No |
| 3 | SESN2 | sestrin 2 [Source:HGNC Symbol;Acc:HGNC:20746] | 1059 | 2.220 | 0.0182 | No |
| 4 | MECR | mitochondrial trans-2-enoyl-CoA reductase [Source:HGNC Symbol;Acc:HGNC:19691] | 2564 | 1.099 | -0.0168 | No |
| 5 | ACOX1 | acyl-CoA oxidase 1 [Source:HGNC Symbol;Acc:HGNC:119] | 2736 | 1.010 | -0.0162 | No |
| 6 | AIG1 | androgen induced 1 [Source:HGNC Symbol;Acc:HGNC:21607] | 3604 | 0.639 | -0.0364 | No |
| 7 | AKR1A1 | aldo-keto reductase family 1 member A1 [Source:HGNC Symbol;Acc:HGNC:380] | 3718 | 0.592 | -0.0363 | No |
| 8 | ABHD2 | abhydrolase domain containing 2, acylglycerol lipase [Source:HGNC Symbol;Acc:HGNC:18717] | 3848 | 0.544 | -0.0370 | No |
| 9 | SLC27A2 | solute carrier family 27 member 2 [Source:HGNC Symbol;Acc:HGNC:10996] | 4412 | 0.468 | -0.0498 | No |
| 10 | PGD | phosphogluconate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:8891] | 4840 | 0.336 | -0.0596 | No |
| 11 | ADIPOQ | adiponectin, C1Q and collagen domain containing [Source:HGNC Symbol;Acc:HGNC:13633] | 5232 | 0.207 | -0.0691 | No |
| 12 | CPT1B | carnitine palmitoyltransferase 1B [Source:HGNC Symbol;Acc:HGNC:2329] | 5435 | 0.149 | -0.0738 | No |
| 13 | ABHD1 | abhydrolase domain containing 1 [Source:HGNC Symbol;Acc:HGNC:17553] | 5830 | 0.037 | -0.0843 | No |
| 14 | EHHADH | enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3247] | 7639 | 0.000 | -0.1332 | No |
| 15 | NUDT7 | nudix hydrolase 7 [Source:HGNC Symbol;Acc:HGNC:8054] | 7869 | 0.000 | -0.1394 | No |
| 16 | GLYATL2 | glycine-N-acyltransferase like 2 [Source:HGNC Symbol;Acc:HGNC:24178] | 10070 | 0.000 | -0.1989 | No |
| 17 | AGXT2 | alanine--glyoxylate aminotransferase 2 [Source:HGNC Symbol;Acc:HGNC:14412] | 10684 | 0.000 | -0.2155 | No |
| 18 | LDHD | lactate dehydrogenase D [Source:HGNC Symbol;Acc:HGNC:19708] | 10883 | 0.000 | -0.2209 | No |
| 19 | AKR1D1 | aldo-keto reductase family 1 member D1 [Source:HGNC Symbol;Acc:HGNC:388] | 11330 | 0.000 | -0.2330 | No |
| 20 | AGXT | alanine--glyoxylate and serine--pyruvate aminotransferase [Source:HGNC Symbol;Acc:HGNC:341] | 11961 | 0.000 | -0.2500 | No |
| 21 | DECR1 | 2,4-dienoyl-CoA reductase 1 [Source:HGNC Symbol;Acc:HGNC:2753] | 12001 | 0.000 | -0.2511 | No |
| 22 | ACOXL | acyl-CoA oxidase like [Source:HGNC Symbol;Acc:HGNC:25621] | 12117 | 0.000 | -0.2542 | No |
| 23 | ACOX2 | acyl-CoA oxidase 2 [Source:HGNC Symbol;Acc:HGNC:120] | 12122 | 0.000 | -0.2543 | No |
| 24 | CNR1 | cannabinoid receptor 1 [Source:HGNC Symbol;Acc:HGNC:2159] | 12131 | 0.000 | -0.2545 | No |
| 25 | PCK1 | phosphoenolpyruvate carboxykinase 1 [Source:HGNC Symbol;Acc:HGNC:8724] | 15273 | 0.000 | -0.3395 | No |
| 26 | CEL | carboxyl ester lipase [Source:HGNC Symbol;Acc:HGNC:1848] | 15668 | 0.000 | -0.3501 | No |
| 27 | PLIN5 | perilipin 5 [Source:HGNC Symbol;Acc:HGNC:33196] | 15705 | 0.000 | -0.3511 | No |
| 28 | BDH2 | 3-hydroxybutyrate dehydrogenase 2 [Source:HGNC Symbol;Acc:HGNC:32389] | 16070 | 0.000 | -0.3610 | No |
| 29 | ABHD3 | abhydrolase domain containing 3, phospholipase [Source:HGNC Symbol;Acc:HGNC:18718] | 16431 | 0.000 | -0.3707 | No |
| 30 | TWIST1 | twist family bHLH transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:12428] | 17199 | 0.000 | -0.3915 | No |
| 31 | ADTRP | androgen dependent TFPI regulating protein [Source:HGNC Symbol;Acc:HGNC:21214] | 18468 | 0.000 | -0.4258 | No |
| 32 | LEP | leptin [Source:HGNC Symbol;Acc:HGNC:6553] | 18528 | 0.000 | -0.4274 | No |
| 33 | ECHDC2 | enoyl-CoA hydratase domain containing 2 [Source:HGNC Symbol;Acc:HGNC:23408] | 19795 | 0.000 | -0.4616 | No |
| 34 | SULT2A1 | sulfotransferase family 2A member 1 [Source:HGNC Symbol;Acc:HGNC:11458] | 20653 | 0.000 | -0.4848 | No |
| 35 | CRABP1 | cellular retinoic acid binding protein 1 [Source:HGNC Symbol;Acc:HGNC:2338] | 20945 | 0.000 | -0.4927 | No |
| 36 | PPARA | peroxisome proliferator activated receptor alpha [Source:HGNC Symbol;Acc:HGNC:9232] | 21005 | 0.000 | -0.4943 | No |
| 37 | CYP4A11 | cytochrome P450 family 4 subfamily A member 11 [Source:HGNC Symbol;Acc:HGNC:2642] | 21316 | 0.000 | -0.5027 | No |
| 38 | HAO2 | hydroxyacid oxidase 2 [Source:HGNC Symbol;Acc:HGNC:4810] | 21850 | 0.000 | -0.5171 | No |
| 39 | ABCB11 | ATP binding cassette subfamily B member 11 [Source:HGNC Symbol;Acc:HGNC:42] | 23470 | 0.000 | -0.5609 | No |
| 40 | IRS1 | insulin receptor substrate 1 [Source:HGNC Symbol;Acc:HGNC:6125] | 26625 | 0.000 | -0.6462 | No |
| 41 | IRS2 | insulin receptor substrate 2 [Source:HGNC Symbol;Acc:HGNC:6126] | 26628 | 0.000 | -0.6463 | No |
| 42 | CYP2W1 | cytochrome P450 family 2 subfamily W member 1 [Source:HGNC Symbol;Acc:HGNC:20243] | 27576 | 0.000 | -0.6719 | No |
| 43 | PEX13 | peroxisomal biogenesis factor 13 [Source:HGNC Symbol;Acc:HGNC:8855] | 28308 | -0.057 | -0.6914 | No |
| 44 | MLYCD | malonyl-CoA decarboxylase [Source:HGNC Symbol;Acc:HGNC:7150] | 28388 | -0.081 | -0.6931 | No |
| 45 | CYP4F11 | cytochrome P450 family 4 subfamily F member 11 [Source:HGNC Symbol;Acc:HGNC:13265] | 28427 | -0.083 | -0.6937 | No |
| 46 | AKT2 | AKT serine/threonine kinase 2 [Source:HGNC Symbol;Acc:HGNC:392] | 28441 | -0.087 | -0.6936 | No |
| 47 | ALDH3A2 | aldehyde dehydrogenase 3 family member A2 [Source:HGNC Symbol;Acc:HGNC:403] | 28466 | -0.093 | -0.6938 | No |
| 48 | ILVBL | ilvB acetolactate synthase like [Source:HGNC Symbol;Acc:HGNC:6041] | 28748 | -0.170 | -0.7005 | No |
| 49 | HADHA | hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha [Source:HGNC Symbol;Acc:HGNC:4801] | 29031 | -0.244 | -0.7069 | No |
| 50 | MMAA | metabolism of cobalamin associated A [Source:HGNC Symbol;Acc:HGNC:18871] | 29412 | -0.344 | -0.7154 | No |
| 51 | CYP26B1 | cytochrome P450 family 26 subfamily B member 1 [Source:HGNC Symbol;Acc:HGNC:20581] | 29563 | -0.376 | -0.7175 | No |
| 52 | ETFA | electron transfer flavoprotein subunit alpha [Source:HGNC Symbol;Acc:HGNC:3481] | 29688 | -0.414 | -0.7187 | No |
| 53 | PEX2 | peroxisomal biogenesis factor 2 [Source:HGNC Symbol;Acc:HGNC:9717] | 29796 | -0.441 | -0.7193 | No |
| 54 | PPARD | peroxisome proliferator activated receptor delta [Source:HGNC Symbol;Acc:HGNC:9235] | 29996 | -0.499 | -0.7221 | No |
| 55 | DECR2 | 2,4-dienoyl-CoA reductase 2 [Source:HGNC Symbol;Acc:HGNC:2754] | 30235 | -0.537 | -0.7257 | No |
| 56 | ACADVL | acyl-CoA dehydrogenase very long chain [Source:HGNC Symbol;Acc:HGNC:92] | 30784 | -0.705 | -0.7369 | No |
| 57 | ACAD11 | acyl-CoA dehydrogenase family member 11 [Source:HGNC Symbol;Acc:HGNC:30211] | 30934 | -0.747 | -0.7371 | Yes |
| 58 | AKT1 | AKT serine/threonine kinase 1 [Source:HGNC Symbol;Acc:HGNC:391] | 30942 | -0.750 | -0.7334 | Yes |
| 59 | AMACR | alpha-methylacyl-CoA racemase [Source:HGNC Symbol;Acc:HGNC:451] | 31043 | -0.783 | -0.7320 | Yes |
| 60 | ACOT8 | acyl-CoA thioesterase 8 [Source:HGNC Symbol;Acc:HGNC:15919] | 31179 | -0.818 | -0.7314 | Yes |
| 61 | MTOR | mechanistic target of rapamycin kinase [Source:HGNC Symbol;Acc:HGNC:3942] | 31224 | -0.836 | -0.7283 | Yes |
| 62 | SLC27A4 | solute carrier family 27 member 4 [Source:HGNC Symbol;Acc:HGNC:10998] | 31634 | -0.968 | -0.7344 | Yes |
| 63 | LPIN2 | lipin 2 [Source:HGNC Symbol;Acc:HGNC:14450] | 31771 | -1.014 | -0.7328 | Yes |
| 64 | CPT1C | carnitine palmitoyltransferase 1C [Source:HGNC Symbol;Acc:HGNC:18540] | 31792 | -1.022 | -0.7280 | Yes |
| 65 | SLC25A17 | solute carrier family 25 member 17 [Source:HGNC Symbol;Acc:HGNC:10987] | 31998 | -1.092 | -0.7279 | Yes |
| 66 | ETFB | electron transfer flavoprotein subunit beta [Source:HGNC Symbol;Acc:HGNC:3482] | 32118 | -1.145 | -0.7252 | Yes |
| 67 | LONP2 | lon peptidase 2, peroxisomal [Source:HGNC Symbol;Acc:HGNC:20598] | 32216 | -1.184 | -0.7217 | Yes |
| 68 | GCDH | glutaryl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4189] | 32553 | -1.248 | -0.7243 | Yes |
| 69 | PCK2 | phosphoenolpyruvate carboxykinase 2, mitochondrial [Source:HGNC Symbol;Acc:HGNC:8725] | 32634 | -1.286 | -0.7198 | Yes |
| 70 | CYP4F2 | cytochrome P450 family 4 subfamily F member 2 [Source:HGNC Symbol;Acc:HGNC:2645] | 32709 | -1.325 | -0.7150 | Yes |
| 71 | ACADL | acyl-CoA dehydrogenase long chain [Source:HGNC Symbol;Acc:HGNC:88] | 32942 | -1.418 | -0.7139 | Yes |
| 72 | ETFDH | electron transfer flavoprotein dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3483] | 33158 | -1.512 | -0.7119 | Yes |
| 73 | FABP1 | fatty acid binding protein 1 [Source:HGNC Symbol;Acc:HGNC:3555] | 33226 | -1.545 | -0.7057 | Yes |
| 74 | ACBD5 | acyl-CoA binding domain containing 5 [Source:HGNC Symbol;Acc:HGNC:23338] | 33272 | -1.562 | -0.6988 | Yes |
| 75 | PEX7 | peroxisomal biogenesis factor 7 [Source:HGNC Symbol;Acc:HGNC:8860] | 33309 | -1.580 | -0.6916 | Yes |
| 76 | CYP4F12 | cytochrome P450 family 4 subfamily F member 12 [Source:HGNC Symbol;Acc:HGNC:18857] | 33338 | -1.587 | -0.6842 | Yes |
| 77 | PHYH | phytanoyl-CoA 2-hydroxylase [Source:HGNC Symbol;Acc:HGNC:8940] | 33345 | -1.592 | -0.6761 | Yes |
| 78 | HADHB | hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta [Source:HGNC Symbol;Acc:HGNC:4803] | 33471 | -1.650 | -0.6709 | Yes |
| 79 | ACAT1 | acetyl-CoA acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:93] | 33726 | -1.764 | -0.6686 | Yes |
| 80 | CYP26A1 | cytochrome P450 family 26 subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:2603] | 33779 | -1.798 | -0.6607 | Yes |
| 81 | NUDT19 | nudix hydrolase 19 [Source:HGNC Symbol;Acc:HGNC:32036] | 33829 | -1.826 | -0.6526 | Yes |
| 82 | ECHDC1 | ethylmalonyl-CoA decarboxylase 1 [Source:HGNC Symbol;Acc:HGNC:21489] | 33912 | -1.872 | -0.6451 | Yes |
| 83 | ECI1 | enoyl-CoA delta isomerase 1 [Source:HGNC Symbol;Acc:HGNC:2703] | 33932 | -1.880 | -0.6359 | Yes |
| 84 | ABCD3 | ATP binding cassette subfamily D member 3 [Source:HGNC Symbol;Acc:HGNC:67] | 34018 | -1.925 | -0.6282 | Yes |
| 85 | IVD | isovaleryl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:6186] | 34142 | -1.990 | -0.6213 | Yes |
| 86 | CPT2 | carnitine palmitoyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:2330] | 34181 | -2.015 | -0.6118 | Yes |
| 87 | PON3 | paraoxonase 3 [Source:HGNC Symbol;Acc:HGNC:9206] | 34280 | -2.066 | -0.6038 | Yes |
| 88 | PCCB | propionyl-CoA carboxylase subunit beta [Source:HGNC Symbol;Acc:HGNC:8654] | 34343 | -2.114 | -0.5945 | Yes |
| 89 | ABCD4 | ATP binding cassette subfamily D member 4 [Source:HGNC Symbol;Acc:HGNC:68] | 34346 | -2.119 | -0.5836 | Yes |
| 90 | IDNK | IDNK gluconokinase [Source:HGNC Symbol;Acc:HGNC:31367] | 34421 | -2.127 | -0.5746 | Yes |
| 91 | HOGA1 | 4-hydroxy-2-oxoglutarate aldolase 1 [Source:HGNC Symbol;Acc:HGNC:25155] | 34463 | -2.130 | -0.5647 | Yes |
| 92 | FAH | fumarylacetoacetate hydrolase [Source:HGNC Symbol;Acc:HGNC:3579] | 34494 | -2.151 | -0.5543 | Yes |
| 93 | ACADM | acyl-CoA dehydrogenase medium chain [Source:HGNC Symbol;Acc:HGNC:89] | 34543 | -2.191 | -0.5443 | Yes |
| 94 | CYP4F3 | cytochrome P450 family 4 subfamily F member 3 [Source:HGNC Symbol;Acc:HGNC:2646] | 34550 | -2.196 | -0.5331 | Yes |
| 95 | DCXR | dicarbonyl and L-xylulose reductase [Source:HGNC Symbol;Acc:HGNC:18985] | 34630 | -2.251 | -0.5236 | Yes |
| 96 | PLA2G15 | phospholipase A2 group XV [Source:HGNC Symbol;Acc:HGNC:17163] | 34637 | -2.255 | -0.5120 | Yes |
| 97 | PECR | peroxisomal trans-2-enoyl-CoA reductase [Source:HGNC Symbol;Acc:HGNC:18281] | 34643 | -2.257 | -0.5005 | Yes |
| 98 | CRAT | carnitine O-acetyltransferase [Source:HGNC Symbol;Acc:HGNC:2342] | 34690 | -2.286 | -0.4899 | Yes |
| 99 | SCP2 | sterol carrier protein 2 [Source:HGNC Symbol;Acc:HGNC:10606] | 34707 | -2.292 | -0.4784 | Yes |
| 100 | ECHS1 | enoyl-CoA hydratase, short chain 1 [Source:HGNC Symbol;Acc:HGNC:3151] | 34764 | -2.324 | -0.4679 | Yes |
| 101 | XYLB | xylulokinase [Source:HGNC Symbol;Acc:HGNC:12839] | 34766 | -2.324 | -0.4559 | Yes |
| 102 | TYSND1 | trypsin domain containing 1 [Source:HGNC Symbol;Acc:HGNC:28531] | 34886 | -2.411 | -0.4466 | Yes |
| 103 | ACAA1 | acetyl-CoA acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:82] | 34902 | -2.428 | -0.4345 | Yes |
| 104 | HSD17B4 | hydroxysteroid 17-beta dehydrogenase 4 [Source:HGNC Symbol;Acc:HGNC:5213] | 34969 | -2.461 | -0.4235 | Yes |
| 105 | ECI2 | enoyl-CoA delta isomerase 2 [Source:HGNC Symbol;Acc:HGNC:14601] | 34973 | -2.463 | -0.4108 | Yes |
| 106 | HSD17B10 | hydroxysteroid 17-beta dehydrogenase 10 [Source:HGNC Symbol;Acc:HGNC:4800] | 35050 | -2.525 | -0.3998 | Yes |
| 107 | MMUT | methylmalonyl-CoA mutase [Source:HGNC Symbol;Acc:HGNC:7526] | 35155 | -2.603 | -0.3892 | Yes |
| 108 | MFSD2A | major facilitator superfamily domain containing 2A [Source:HGNC Symbol;Acc:HGNC:25897] | 35164 | -2.610 | -0.3759 | Yes |
| 109 | MCEE | methylmalonyl-CoA epimerase [Source:HGNC Symbol;Acc:HGNC:16732] | 35430 | -2.779 | -0.3686 | Yes |
| 110 | CPT1A | carnitine palmitoyltransferase 1A [Source:HGNC Symbol;Acc:HGNC:2328] | 35438 | -2.788 | -0.3544 | Yes |
| 111 | AUH | AU RNA binding methylglutaconyl-CoA hydratase [Source:HGNC Symbol;Acc:HGNC:890] | 35516 | -2.861 | -0.3416 | Yes |
| 112 | MCAT | malonyl-CoA-acyl carrier protein transacylase [Source:HGNC Symbol;Acc:HGNC:29622] | 35728 | -3.062 | -0.3315 | Yes |
| 113 | ACADS | acyl-CoA dehydrogenase short chain [Source:HGNC Symbol;Acc:HGNC:90] | 35735 | -3.069 | -0.3158 | Yes |
| 114 | ACAT2 | acetyl-CoA acetyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:94] | 35890 | -3.248 | -0.3031 | Yes |
| 115 | FAAH | fatty acid amide hydrolase [Source:HGNC Symbol;Acc:HGNC:3553] | 35895 | -3.253 | -0.2864 | Yes |
| 116 | ACAD10 | acyl-CoA dehydrogenase family member 10 [Source:HGNC Symbol;Acc:HGNC:21597] | 35907 | -3.267 | -0.2697 | Yes |
| 117 | PCCA | propionyl-CoA carboxylase subunit alpha [Source:HGNC Symbol;Acc:HGNC:8653] | 35937 | -3.301 | -0.2534 | Yes |
| 118 | LPIN3 | lipin 3 [Source:HGNC Symbol;Acc:HGNC:14451] | 36036 | -3.371 | -0.2386 | Yes |
| 119 | ACACB | acetyl-CoA carboxylase beta [Source:HGNC Symbol;Acc:HGNC:85] | 36184 | -3.516 | -0.2244 | Yes |
| 120 | CRYL1 | crystallin lambda 1 [Source:HGNC Symbol;Acc:HGNC:18246] | 36250 | -3.579 | -0.2076 | Yes |
| 121 | ECH1 | enoyl-CoA hydratase 1 [Source:HGNC Symbol;Acc:HGNC:3149] | 36319 | -3.655 | -0.1905 | Yes |
| 122 | ACAA2 | acetyl-CoA acyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:83] | 36335 | -3.680 | -0.1719 | Yes |
| 123 | SORD | sorbitol dehydrogenase [Source:HGNC Symbol;Acc:HGNC:11184] | 36402 | -3.755 | -0.1542 | Yes |
| 124 | HADH | hydroxyacyl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4799] | 36412 | -3.775 | -0.1349 | Yes |
| 125 | ABCD1 | ATP binding cassette subfamily D member 1 [Source:HGNC Symbol;Acc:HGNC:61] | 36470 | -3.832 | -0.1166 | Yes |
| 126 | ETFBKMT | electron transfer flavoprotein subunit beta lysine methyltransferase [Source:HGNC Symbol;Acc:HGNC:28739] | 36586 | -3.990 | -0.0990 | Yes |
| 127 | CROT | carnitine O-octanoyltransferase [Source:HGNC Symbol;Acc:HGNC:2366] | 36628 | -4.035 | -0.0793 | Yes |
| 128 | ACOX3 | acyl-CoA oxidase 3, pristanoyl [Source:HGNC Symbol;Acc:HGNC:121] | 36791 | -4.254 | -0.0616 | Yes |
| 129 | HACL1 | 2-hydroxyacyl-CoA lyase 1 [Source:HGNC Symbol;Acc:HGNC:17856] | 36850 | -4.371 | -0.0405 | Yes |
| 130 | ABCD2 | ATP binding cassette subfamily D member 2 [Source:HGNC Symbol;Acc:HGNC:66] | 36913 | -4.479 | -0.0190 | Yes |
| 131 | LPIN1 | lipin 1 [Source:HGNC Symbol;Acc:HGNC:13345] | 36962 | -4.592 | 0.0035 | Yes |