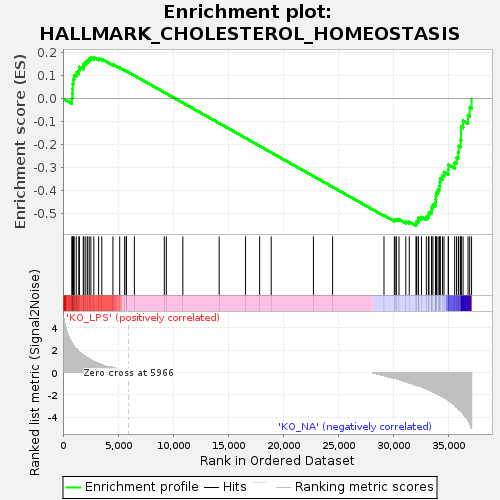
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.55378795 |
| Normalized Enrichment Score (NES) | -1.1067101 |
| Nominal p-value | 0.29418743 |
| FDR q-value | 0.5143366 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ACTG1 | actin gamma 1 [Source:HGNC Symbol;Acc:HGNC:144] | 802 | 2.641 | 0.0010 | No |
| 2 | CHKA | choline kinase alpha [Source:HGNC Symbol;Acc:HGNC:1937] | 853 | 2.554 | 0.0216 | No |
| 3 | ERRFI1 | ERBB receptor feedback inhibitor 1 [Source:HGNC Symbol;Acc:HGNC:18185] | 864 | 2.531 | 0.0430 | No |
| 4 | S100A11 | S100 calcium binding protein A11 [Source:HGNC Symbol;Acc:HGNC:10488] | 890 | 2.512 | 0.0639 | No |
| 5 | ATF3 | activating transcription factor 3 [Source:HGNC Symbol;Acc:HGNC:785] | 947 | 2.411 | 0.0831 | No |
| 6 | ATF5 | activating transcription factor 5 [Source:HGNC Symbol;Acc:HGNC:790] | 1017 | 2.290 | 0.1009 | No |
| 7 | HSD17B7 | hydroxysteroid 17-beta dehydrogenase 7 [Source:HGNC Symbol;Acc:HGNC:5215] | 1202 | 2.075 | 0.1137 | No |
| 8 | ANTXR2 | ANTXR cell adhesion molecule 2 [Source:HGNC Symbol;Acc:HGNC:21732] | 1434 | 1.870 | 0.1235 | No |
| 9 | ALCAM | activated leukocyte cell adhesion molecule [Source:HGNC Symbol;Acc:HGNC:400] | 1490 | 1.822 | 0.1377 | No |
| 10 | HMGCR | 3-hydroxy-3-methylglutaryl-CoA reductase [Source:HGNC Symbol;Acc:HGNC:5006] | 1841 | 1.517 | 0.1413 | No |
| 11 | TRIB3 | tribbles pseudokinase 3 [Source:HGNC Symbol;Acc:HGNC:16228] | 1899 | 1.465 | 0.1523 | No |
| 12 | PLSCR1 | phospholipid scramblase 1 [Source:HGNC Symbol;Acc:HGNC:9092] | 2072 | 1.379 | 0.1595 | No |
| 13 | GNAI1 | G protein subunit alpha i1 [Source:HGNC Symbol;Acc:HGNC:4384] | 2237 | 1.283 | 0.1661 | No |
| 14 | FABP5 | fatty acid binding protein 5 [Source:HGNC Symbol;Acc:HGNC:3560] | 2381 | 1.201 | 0.1725 | No |
| 15 | CD9 | CD9 molecule [Source:HGNC Symbol;Acc:HGNC:1709] | 2531 | 1.123 | 0.1781 | No |
| 16 | TNFRSF12A | TNF receptor superfamily member 12A [Source:HGNC Symbol;Acc:HGNC:18152] | 2798 | 0.981 | 0.1794 | No |
| 17 | ANXA5 | annexin A5 [Source:HGNC Symbol;Acc:HGNC:543] | 3233 | 0.783 | 0.1744 | No |
| 18 | LDLR | low density lipoprotein receptor [Source:HGNC Symbol;Acc:HGNC:6547] | 3521 | 0.673 | 0.1724 | No |
| 19 | CTNNB1 | catenin beta 1 [Source:HGNC Symbol;Acc:HGNC:2514] | 4531 | 0.430 | 0.1488 | No |
| 20 | STX5 | syntaxin 5 [Source:HGNC Symbol;Acc:HGNC:11440] | 5147 | 0.231 | 0.1342 | No |
| 21 | LPL | lipoprotein lipase [Source:HGNC Symbol;Acc:HGNC:6677] | 5597 | 0.111 | 0.1230 | No |
| 22 | NIBAN1 | niban apoptosis regulator 1 [Source:HGNC Symbol;Acc:HGNC:16784] | 5743 | 0.067 | 0.1197 | No |
| 23 | IDI1 | isopentenyl-diphosphate delta isomerase 1 [Source:HGNC Symbol;Acc:HGNC:5387] | 5754 | 0.064 | 0.1199 | No |
| 24 | FBXO6 | F-box protein 6 [Source:HGNC Symbol;Acc:HGNC:13585] | 6477 | 0.000 | 0.1004 | No |
| 25 | GPX8 | glutathione peroxidase 8 (putative) [Source:HGNC Symbol;Acc:HGNC:33100] | 9198 | 0.000 | 0.0270 | No |
| 26 | GSTM2 | glutathione S-transferase mu 2 [Source:HGNC Symbol;Acc:HGNC:4634] | 9379 | 0.000 | 0.0221 | No |
| 27 | ADH4 | alcohol dehydrogenase 4 (class II), pi polypeptide [Source:HGNC Symbol;Acc:HGNC:252] | 10880 | 0.000 | -0.0184 | No |
| 28 | ANXA13 | annexin A13 [Source:HGNC Symbol;Acc:HGNC:536] | 14176 | 0.000 | -0.1074 | No |
| 29 | AVPR1A | arginine vasopressin receptor 1A [Source:HGNC Symbol;Acc:HGNC:895] | 16571 | 0.000 | -0.1721 | No |
| 30 | SEMA3B | semaphorin 3B [Source:HGNC Symbol;Acc:HGNC:10724] | 17850 | 0.000 | -0.2066 | No |
| 31 | CXCL16 | C-X-C motif chemokine ligand 16 [Source:HGNC Symbol;Acc:HGNC:16642] | 18899 | 0.000 | -0.2350 | No |
| 32 | PLAUR | plasminogen activator, urokinase receptor [Source:HGNC Symbol;Acc:HGNC:9053] | 22732 | 0.000 | -0.3385 | No |
| 33 | MAL2 | mal, T cell differentiation protein 2 (gene/pseudogene) [Source:HGNC Symbol;Acc:HGNC:13634] | 24475 | 0.000 | -0.3855 | No |
| 34 | NFIL3 | nuclear factor, interleukin 3 regulated [Source:HGNC Symbol;Acc:HGNC:7787] | 29139 | -0.266 | -0.5092 | No |
| 35 | FASN | fatty acid synthase [Source:HGNC Symbol;Acc:HGNC:3594] | 30079 | -0.520 | -0.5301 | No |
| 36 | PPARG | peroxisome proliferator activated receptor gamma [Source:HGNC Symbol;Acc:HGNC:9236] | 30143 | -0.537 | -0.5272 | No |
| 37 | CPEB2 | cytoplasmic polyadenylation element binding protein 2 [Source:HGNC Symbol;Acc:HGNC:21745] | 30265 | -0.544 | -0.5258 | No |
| 38 | GUSB | glucuronidase beta [Source:HGNC Symbol;Acc:HGNC:4696] | 30500 | -0.624 | -0.5268 | No |
| 39 | EBP | EBP cholestenol delta-isomerase [Source:HGNC Symbol;Acc:HGNC:3133] | 31120 | -0.796 | -0.5367 | No |
| 40 | JAG1 | jagged canonical Notch ligand 1 [Source:HGNC Symbol;Acc:HGNC:6188] | 31434 | -0.906 | -0.5374 | No |
| 41 | SREBF2 | sterol regulatory element binding transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:11290] | 32043 | -1.110 | -0.5443 | Yes |
| 42 | CLU | clusterin [Source:HGNC Symbol;Acc:HGNC:2095] | 32096 | -1.134 | -0.5359 | Yes |
| 43 | HMGCS1 | 3-hydroxy-3-methylglutaryl-CoA synthase 1 [Source:HGNC Symbol;Acc:HGNC:5007] | 32257 | -1.201 | -0.5299 | Yes |
| 44 | CYP51A1 | cytochrome P450 family 51 subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:2649] | 32260 | -1.203 | -0.5197 | Yes |
| 45 | MVK | mevalonate kinase [Source:HGNC Symbol;Acc:HGNC:7530] | 32540 | -1.241 | -0.5166 | Yes |
| 46 | PDK3 | pyruvate dehydrogenase kinase 3 [Source:HGNC Symbol;Acc:HGNC:8811] | 32983 | -1.443 | -0.5161 | Yes |
| 47 | STARD4 | StAR related lipid transfer domain containing 4 [Source:HGNC Symbol;Acc:HGNC:18058] | 33170 | -1.516 | -0.5081 | Yes |
| 48 | GLDC | glycine decarboxylase [Source:HGNC Symbol;Acc:HGNC:4313] | 33222 | -1.543 | -0.4963 | Yes |
| 49 | ATXN2 | ataxin 2 [Source:HGNC Symbol;Acc:HGNC:10555] | 33453 | -1.644 | -0.4884 | Yes |
| 50 | SC5D | sterol-C5-desaturase [Source:HGNC Symbol;Acc:HGNC:10547] | 33472 | -1.651 | -0.4747 | Yes |
| 51 | LGALS3 | galectin 3 [Source:HGNC Symbol;Acc:HGNC:6563] | 33581 | -1.709 | -0.4629 | Yes |
| 52 | PNRC1 | proline rich nuclear receptor coactivator 1 [Source:HGNC Symbol;Acc:HGNC:17278] | 33809 | -1.816 | -0.4535 | Yes |
| 53 | SQLE | squalene epoxidase [Source:HGNC Symbol;Acc:HGNC:11279] | 33840 | -1.829 | -0.4386 | Yes |
| 54 | FDFT1 | farnesyl-diphosphate farnesyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:3629] | 33860 | -1.844 | -0.4233 | Yes |
| 55 | TMEM97 | transmembrane protein 97 [Source:HGNC Symbol;Acc:HGNC:28106] | 33898 | -1.867 | -0.4082 | Yes |
| 56 | PMVK | phosphomevalonate kinase [Source:HGNC Symbol;Acc:HGNC:9141] | 34046 | -1.938 | -0.3956 | Yes |
| 57 | FDPS | farnesyl diphosphate synthase [Source:HGNC Symbol;Acc:HGNC:3631] | 34167 | -2.006 | -0.3816 | Yes |
| 58 | MVD | mevalonate diphosphate decarboxylase [Source:HGNC Symbol;Acc:HGNC:7529] | 34212 | -2.031 | -0.3653 | Yes |
| 59 | TM7SF2 | transmembrane 7 superfamily member 2 [Source:HGNC Symbol;Acc:HGNC:11863] | 34251 | -2.052 | -0.3488 | Yes |
| 60 | NSDHL | NAD(P) dependent steroid dehydrogenase-like [Source:HGNC Symbol;Acc:HGNC:13398] | 34450 | -2.127 | -0.3359 | Yes |
| 61 | LSS | lanosterol synthase [Source:HGNC Symbol;Acc:HGNC:6708] | 34586 | -2.224 | -0.3204 | Yes |
| 62 | ALDOC | aldolase, fructose-bisphosphate C [Source:HGNC Symbol;Acc:HGNC:418] | 34979 | -2.470 | -0.3098 | Yes |
| 63 | PCYT2 | phosphate cytidylyltransferase 2, ethanolamine [Source:HGNC Symbol;Acc:HGNC:8756] | 34982 | -2.474 | -0.2886 | Yes |
| 64 | DHCR7 | 7-dehydrocholesterol reductase [Source:HGNC Symbol;Acc:HGNC:2860] | 35557 | -2.893 | -0.2793 | Yes |
| 65 | ACSS2 | acyl-CoA synthetase short chain family member 2 [Source:HGNC Symbol;Acc:HGNC:15814] | 35729 | -3.064 | -0.2576 | Yes |
| 66 | ACAT2 | acetyl-CoA acetyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:94] | 35890 | -3.248 | -0.2341 | Yes |
| 67 | ABCA2 | ATP binding cassette subfamily A member 2 [Source:HGNC Symbol;Acc:HGNC:32] | 35928 | -3.289 | -0.2068 | Yes |
| 68 | TP53INP1 | tumor protein p53 inducible nuclear protein 1 [Source:HGNC Symbol;Acc:HGNC:18022] | 36095 | -3.432 | -0.1819 | Yes |
| 69 | LGMN | legumain [Source:HGNC Symbol;Acc:HGNC:9472] | 36139 | -3.470 | -0.1532 | Yes |
| 70 | FADS2 | fatty acid desaturase 2 [Source:HGNC Symbol;Acc:HGNC:3575] | 36140 | -3.471 | -0.1234 | Yes |
| 71 | ECH1 | enoyl-CoA hydratase 1 [Source:HGNC Symbol;Acc:HGNC:3149] | 36319 | -3.655 | -0.0969 | Yes |
| 72 | ETHE1 | ETHE1 persulfide dioxygenase [Source:HGNC Symbol;Acc:HGNC:23287] | 36769 | -4.215 | -0.0728 | Yes |
| 73 | SCD | stearoyl-CoA desaturase [Source:HGNC Symbol;Acc:HGNC:10571] | 36921 | -4.491 | -0.0384 | Yes |
| 74 | CBS | cystathionine beta-synthase [Source:HGNC Symbol;Acc:HGNC:1550] | 37090 | -5.000 | 0.0000 | Yes |