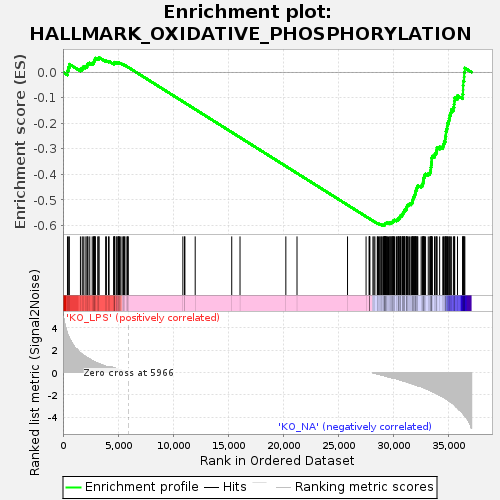
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HALLMARK_OXIDATIVE_PHOSPHORYLATION |
| Enrichment Score (ES) | -0.60066444 |
| Normalized Enrichment Score (NES) | -1.3017744 |
| Nominal p-value | 0.0135557875 |
| FDR q-value | 0.18086132 |
| FWER p-Value | 0.734 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SDHD | succinate dehydrogenase complex subunit D [Source:HGNC Symbol;Acc:HGNC:10683] | 406 | 3.441 | 0.0056 | No |
| 2 | POR | cytochrome p450 oxidoreductase [Source:HGNC Symbol;Acc:HGNC:9208] | 483 | 3.259 | 0.0194 | No |
| 3 | COX17 | cytochrome c oxidase copper chaperone COX17 [Source:HGNC Symbol;Acc:HGNC:2264] | 577 | 3.043 | 0.0316 | No |
| 4 | TCIRG1 | T cell immune regulator 1, ATPase H+ transporting V0 subunit a3 [Source:HGNC Symbol;Acc:HGNC:11647] | 1583 | 1.731 | 0.0127 | No |
| 5 | CASP7 | caspase 7 [Source:HGNC Symbol;Acc:HGNC:1508] | 1743 | 1.591 | 0.0161 | No |
| 6 | ATP6V1E1 | ATPase H+ transporting V1 subunit E1 [Source:HGNC Symbol;Acc:HGNC:857] | 1838 | 1.521 | 0.0209 | No |
| 7 | CYCS | cytochrome c, somatic [Source:HGNC Symbol;Acc:HGNC:19986] | 2006 | 1.426 | 0.0233 | No |
| 8 | TIMM10 | translocase of inner mitochondrial membrane 10 [Source:HGNC Symbol;Acc:HGNC:11814] | 2175 | 1.314 | 0.0251 | No |
| 9 | ATP6V1H | ATPase H+ transporting V1 subunit H [Source:HGNC Symbol;Acc:HGNC:18303] | 2198 | 1.296 | 0.0308 | No |
| 10 | TIMM17A | translocase of inner mitochondrial membrane 17A [Source:HGNC Symbol;Acc:HGNC:17315] | 2270 | 1.263 | 0.0349 | No |
| 11 | UQCR11 | ubiquinol-cytochrome c reductase, complex III subunit XI [Source:HGNC Symbol;Acc:HGNC:30862] | 2429 | 1.172 | 0.0363 | No |
| 12 | PRDX3 | peroxiredoxin 3 [Source:HGNC Symbol;Acc:HGNC:9354] | 2652 | 1.047 | 0.0354 | No |
| 13 | UQCRB | ubiquinol-cytochrome c reductase binding protein [Source:HGNC Symbol;Acc:HGNC:12582] | 2768 | 0.994 | 0.0371 | No |
| 14 | ALAS1 | 5'-aminolevulinate synthase 1 [Source:HGNC Symbol;Acc:HGNC:396] | 2802 | 0.977 | 0.0409 | No |
| 15 | DLD | dihydrolipoamide dehydrogenase [Source:HGNC Symbol;Acc:HGNC:2898] | 2834 | 0.961 | 0.0447 | No |
| 16 | COX15 | cytochrome c oxidase assembly homolog COX15 [Source:HGNC Symbol;Acc:HGNC:2263] | 2873 | 0.937 | 0.0482 | No |
| 17 | MRPL35 | mitochondrial ribosomal protein L35 [Source:HGNC Symbol;Acc:HGNC:14489] | 2900 | 0.926 | 0.0520 | No |
| 18 | MFN2 | mitofusin 2 [Source:HGNC Symbol;Acc:HGNC:16877] | 2939 | 0.910 | 0.0554 | No |
| 19 | SLC25A12 | solute carrier family 25 member 12 [Source:HGNC Symbol;Acc:HGNC:10982] | 3153 | 0.816 | 0.0535 | No |
| 20 | SLC25A20 | solute carrier family 25 member 20 [Source:HGNC Symbol;Acc:HGNC:1421] | 3256 | 0.776 | 0.0545 | No |
| 21 | UQCRQ | ubiquinol-cytochrome c reductase complex III subunit VII [Source:HGNC Symbol;Acc:HGNC:29594] | 3264 | 0.772 | 0.0581 | No |
| 22 | TIMM9 | translocase of inner mitochondrial membrane 9 [Source:HGNC Symbol;Acc:HGNC:11819] | 3871 | 0.535 | 0.0442 | No |
| 23 | ATP6V1C1 | ATPase H+ transporting V1 subunit C1 [Source:HGNC Symbol;Acc:HGNC:856] | 3941 | 0.510 | 0.0448 | No |
| 24 | NDUFA3 | NADH:ubiquinone oxidoreductase subunit A3 [Source:HGNC Symbol;Acc:HGNC:7686] | 4127 | 0.473 | 0.0421 | No |
| 25 | NDUFB3 | NADH:ubiquinone oxidoreductase subunit B3 [Source:HGNC Symbol;Acc:HGNC:7698] | 4185 | 0.473 | 0.0429 | No |
| 26 | MRPS22 | mitochondrial ribosomal protein S22 [Source:HGNC Symbol;Acc:HGNC:14508] | 4596 | 0.410 | 0.0337 | No |
| 27 | HCCS | holocytochrome c synthase [Source:HGNC Symbol;Acc:HGNC:4837] | 4649 | 0.392 | 0.0342 | No |
| 28 | HSPA9 | heat shock protein family A (Hsp70) member 9 [Source:HGNC Symbol;Acc:HGNC:5244] | 4650 | 0.391 | 0.0361 | No |
| 29 | MRPL11 | mitochondrial ribosomal protein L11 [Source:HGNC Symbol;Acc:HGNC:14042] | 4671 | 0.383 | 0.0374 | No |
| 30 | COX11 | cytochrome c oxidase copper chaperone COX11 [Source:HGNC Symbol;Acc:HGNC:2261] | 4673 | 0.382 | 0.0392 | No |
| 31 | ATP5MG | ATP synthase membrane subunit g [Source:HGNC Symbol;Acc:HGNC:14247] | 4821 | 0.342 | 0.0369 | No |
| 32 | MTX2 | metaxin 2 [Source:HGNC Symbol;Acc:HGNC:7506] | 4886 | 0.321 | 0.0367 | No |
| 33 | NDUFB2 | NADH:ubiquinone oxidoreductase subunit B2 [Source:HGNC Symbol;Acc:HGNC:7697] | 4977 | 0.289 | 0.0357 | No |
| 34 | TIMM50 | translocase of inner mitochondrial membrane 50 [Source:HGNC Symbol;Acc:HGNC:23656] | 5033 | 0.266 | 0.0355 | No |
| 35 | ATP6V0E1 | ATPase H+ transporting V0 subunit e1 [Source:HGNC Symbol;Acc:HGNC:863] | 5039 | 0.263 | 0.0366 | No |
| 36 | NDUFB6 | NADH:ubiquinone oxidoreductase subunit B6 [Source:HGNC Symbol;Acc:HGNC:7701] | 5116 | 0.238 | 0.0357 | No |
| 37 | ATP6V0B | ATPase H+ transporting V0 subunit b [Source:HGNC Symbol;Acc:HGNC:861] | 5118 | 0.237 | 0.0368 | No |
| 38 | ATP6V0C | ATPase H+ transporting V0 subunit c [Source:HGNC Symbol;Acc:HGNC:855] | 5240 | 0.205 | 0.0346 | No |
| 39 | COX6B1 | cytochrome c oxidase subunit 6B1 [Source:HGNC Symbol;Acc:HGNC:2280] | 5371 | 0.168 | 0.0318 | No |
| 40 | TOMM22 | translocase of outer mitochondrial membrane 22 [Source:HGNC Symbol;Acc:HGNC:18002] | 5490 | 0.136 | 0.0293 | No |
| 41 | NDUFS4 | NADH:ubiquinone oxidoreductase subunit S4 [Source:HGNC Symbol;Acc:HGNC:7711] | 5534 | 0.126 | 0.0287 | No |
| 42 | COX7C | cytochrome c oxidase subunit 7C [Source:HGNC Symbol;Acc:HGNC:2292] | 5589 | 0.112 | 0.0278 | No |
| 43 | COX7B | cytochrome c oxidase subunit 7B [Source:HGNC Symbol;Acc:HGNC:2291] | 5727 | 0.072 | 0.0245 | No |
| 44 | UQCR10 | ubiquinol-cytochrome c reductase, complex III subunit X [Source:HGNC Symbol;Acc:HGNC:30863] | 5868 | 0.022 | 0.0208 | No |
| 45 | ATP5F1B | ATP synthase F1 subunit beta [Source:HGNC Symbol;Acc:HGNC:830] | 5873 | 0.020 | 0.0208 | No |
| 46 | GRPEL1 | GrpE like 1, mitochondrial [Source:HGNC Symbol;Acc:HGNC:19696] | 5922 | 0.010 | 0.0195 | No |
| 47 | LDHB | lactate dehydrogenase B [Source:HGNC Symbol;Acc:HGNC:6541] | 10881 | 0.000 | -0.1149 | No |
| 48 | ATP5PO | ATP synthase peripheral stalk subunit OSCP [Source:HGNC Symbol;Acc:HGNC:850] | 11036 | 0.000 | -0.1191 | No |
| 49 | ATP5ME | ATP synthase membrane subunit e [Source:HGNC Symbol;Acc:HGNC:846] | 11062 | 0.000 | -0.1197 | No |
| 50 | DECR1 | 2,4-dienoyl-CoA reductase 1 [Source:HGNC Symbol;Acc:HGNC:2753] | 12001 | 0.000 | -0.1452 | No |
| 51 | COX6C | cytochrome c oxidase subunit 6C [Source:HGNC Symbol;Acc:HGNC:2285] | 15317 | 0.000 | -0.2350 | No |
| 52 | BDH2 | 3-hydroxybutyrate dehydrogenase 2 [Source:HGNC Symbol;Acc:HGNC:32389] | 16070 | 0.000 | -0.2554 | No |
| 53 | ATP1B1 | ATPase Na+/K+ transporting subunit beta 1 [Source:HGNC Symbol;Acc:HGNC:804] | 20229 | 0.000 | -0.3681 | No |
| 54 | COX6A1 | cytochrome c oxidase subunit 6A1 [Source:HGNC Symbol;Acc:HGNC:2277] | 21243 | 0.000 | -0.3956 | No |
| 55 | PDK4 | pyruvate dehydrogenase kinase 4 [Source:HGNC Symbol;Acc:HGNC:8812] | 25829 | 0.000 | -0.5199 | No |
| 56 | ISCA1 | iron-sulfur cluster assembly 1 [Source:HGNC Symbol;Acc:HGNC:28660] | 27512 | 0.000 | -0.5655 | No |
| 57 | NDUFB5 | NADH:ubiquinone oxidoreductase subunit B5 [Source:HGNC Symbol;Acc:HGNC:7700] | 27805 | 0.000 | -0.5734 | No |
| 58 | NDUFB4 | NADH:ubiquinone oxidoreductase subunit B4 [Source:HGNC Symbol;Acc:HGNC:7699] | 27807 | 0.000 | -0.5734 | No |
| 59 | NDUFB1 | NADH:ubiquinone oxidoreductase subunit B1 [Source:HGNC Symbol;Acc:HGNC:7695] | 27812 | 0.000 | -0.5735 | No |
| 60 | NDUFA5 | NADH:ubiquinone oxidoreductase subunit A5 [Source:HGNC Symbol;Acc:HGNC:7688] | 27836 | 0.000 | -0.5741 | No |
| 61 | NDUFA4 | NDUFA4 mitochondrial complex associated [Source:HGNC Symbol;Acc:HGNC:7687] | 27837 | 0.000 | -0.5741 | No |
| 62 | NDUFA1 | NADH:ubiquinone oxidoreductase subunit A1 [Source:HGNC Symbol;Acc:HGNC:7683] | 27841 | 0.000 | -0.5742 | No |
| 63 | MRPS12 | mitochondrial ribosomal protein S12 [Source:HGNC Symbol;Acc:HGNC:10380] | 28135 | -0.006 | -0.5821 | No |
| 64 | COX7A2 | cytochrome c oxidase subunit 7A2 [Source:HGNC Symbol;Acc:HGNC:2288] | 28225 | -0.032 | -0.5844 | No |
| 65 | ATP5MC3 | ATP synthase membrane subunit c locus 3 [Source:HGNC Symbol;Acc:HGNC:843] | 28314 | -0.059 | -0.5865 | No |
| 66 | ATP6V1D | ATPase H+ transporting V1 subunit D [Source:HGNC Symbol;Acc:HGNC:13527] | 28531 | -0.112 | -0.5918 | No |
| 67 | NDUFAB1 | NADH:ubiquinone oxidoreductase subunit AB1 [Source:HGNC Symbol;Acc:HGNC:7694] | 28551 | -0.118 | -0.5917 | No |
| 68 | ATP5PF | ATP synthase peripheral stalk subunit F6 [Source:HGNC Symbol;Acc:HGNC:847] | 28644 | -0.144 | -0.5935 | No |
| 69 | UQCRC2 | ubiquinol-cytochrome c reductase core protein 2 [Source:HGNC Symbol;Acc:HGNC:12586] | 28712 | -0.156 | -0.5946 | No |
| 70 | NDUFA9 | NADH:ubiquinone oxidoreductase subunit A9 [Source:HGNC Symbol;Acc:HGNC:7693] | 28788 | -0.181 | -0.5958 | No |
| 71 | ATP6V1G1 | ATPase H+ transporting V1 subunit G1 [Source:HGNC Symbol;Acc:HGNC:864] | 28871 | -0.199 | -0.5970 | No |
| 72 | MTRR | 5-methyltetrahydrofolate-homocysteine methyltransferase reductase [Source:HGNC Symbol;Acc:HGNC:7473] | 28928 | -0.212 | -0.5975 | No |
| 73 | HADHA | hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha [Source:HGNC Symbol;Acc:HGNC:4801] | 29031 | -0.244 | -0.5991 | No |
| 74 | COX4I1 | cytochrome c oxidase subunit 4I1 [Source:HGNC Symbol;Acc:HGNC:2265] | 29090 | -0.251 | -0.5994 | Yes |
| 75 | PDHB | pyruvate dehydrogenase E1 subunit beta [Source:HGNC Symbol;Acc:HGNC:8808] | 29135 | -0.263 | -0.5994 | Yes |
| 76 | ATP6V1F | ATPase H+ transporting V1 subunit F [Source:HGNC Symbol;Acc:HGNC:16832] | 29150 | -0.267 | -0.5985 | Yes |
| 77 | COX8A | cytochrome c oxidase subunit 8A [Source:HGNC Symbol;Acc:HGNC:2294] | 29195 | -0.282 | -0.5983 | Yes |
| 78 | PMPCA | peptidase, mitochondrial processing subunit alpha [Source:HGNC Symbol;Acc:HGNC:18667] | 29197 | -0.282 | -0.5969 | Yes |
| 79 | VDAC2 | voltage dependent anion channel 2 [Source:HGNC Symbol;Acc:HGNC:12672] | 29212 | -0.287 | -0.5959 | Yes |
| 80 | DLST | dihydrolipoamide S-succinyltransferase [Source:HGNC Symbol;Acc:HGNC:2911] | 29220 | -0.290 | -0.5947 | Yes |
| 81 | SLC25A5 | solute carrier family 25 member 5 [Source:HGNC Symbol;Acc:HGNC:10991] | 29258 | -0.300 | -0.5943 | Yes |
| 82 | IMMT | inner membrane mitochondrial protein [Source:HGNC Symbol;Acc:HGNC:6047] | 29259 | -0.300 | -0.5928 | Yes |
| 83 | SLC25A3 | solute carrier family 25 member 3 [Source:HGNC Symbol;Acc:HGNC:10989] | 29276 | -0.306 | -0.5918 | Yes |
| 84 | NDUFS6 | NADH:ubiquinone oxidoreductase subunit S6 [Source:HGNC Symbol;Acc:HGNC:7713] | 29319 | -0.320 | -0.5914 | Yes |
| 85 | DLAT | dihydrolipoamide S-acetyltransferase [Source:HGNC Symbol;Acc:HGNC:2896] | 29378 | -0.333 | -0.5913 | Yes |
| 86 | ATP5PD | ATP synthase peripheral stalk subunit d [Source:HGNC Symbol;Acc:HGNC:845] | 29397 | -0.338 | -0.5902 | Yes |
| 87 | ATP5F1A | ATP synthase F1 subunit alpha [Source:HGNC Symbol;Acc:HGNC:823] | 29455 | -0.355 | -0.5900 | Yes |
| 88 | NDUFA2 | NADH:ubiquinone oxidoreductase subunit A2 [Source:HGNC Symbol;Acc:HGNC:7685] | 29549 | -0.374 | -0.5907 | Yes |
| 89 | SUCLA2 | succinate-CoA ligase ADP-forming subunit beta [Source:HGNC Symbol;Acc:HGNC:11448] | 29668 | -0.408 | -0.5919 | Yes |
| 90 | ETFA | electron transfer flavoprotein subunit alpha [Source:HGNC Symbol;Acc:HGNC:3481] | 29688 | -0.414 | -0.5905 | Yes |
| 91 | MRPL15 | mitochondrial ribosomal protein L15 [Source:HGNC Symbol;Acc:HGNC:14054] | 29690 | -0.415 | -0.5885 | Yes |
| 92 | CYC1 | cytochrome c1 [Source:HGNC Symbol;Acc:HGNC:2579] | 29789 | -0.439 | -0.5890 | Yes |
| 93 | ACO2 | aconitase 2 [Source:HGNC Symbol;Acc:HGNC:118] | 29849 | -0.455 | -0.5884 | Yes |
| 94 | ACADSB | acyl-CoA dehydrogenase short/branched chain [Source:HGNC Symbol;Acc:HGNC:91] | 29885 | -0.464 | -0.5871 | Yes |
| 95 | MDH2 | malate dehydrogenase 2 [Source:HGNC Symbol;Acc:HGNC:6971] | 29945 | -0.482 | -0.5864 | Yes |
| 96 | TOMM70 | translocase of outer mitochondrial membrane 70 [Source:HGNC Symbol;Acc:HGNC:11985] | 29950 | -0.483 | -0.5841 | Yes |
| 97 | MRPS11 | mitochondrial ribosomal protein S11 [Source:HGNC Symbol;Acc:HGNC:14050] | 29982 | -0.495 | -0.5826 | Yes |
| 98 | ATP5PB | ATP synthase peripheral stalk-membrane subunit b [Source:HGNC Symbol;Acc:HGNC:840] | 30003 | -0.502 | -0.5807 | Yes |
| 99 | ATP5MC1 | ATP synthase membrane subunit c locus 1 [Source:HGNC Symbol;Acc:HGNC:841] | 30098 | -0.527 | -0.5807 | Yes |
| 100 | SUCLG1 | succinate-CoA ligase GDP/ADP-forming subunit alpha [Source:HGNC Symbol;Acc:HGNC:11449] | 30123 | -0.533 | -0.5788 | Yes |
| 101 | AIFM1 | apoptosis inducing factor mitochondria associated 1 [Source:HGNC Symbol;Acc:HGNC:8768] | 30278 | -0.548 | -0.5803 | Yes |
| 102 | ATP5F1E | ATP synthase F1 subunit epsilon [Source:HGNC Symbol;Acc:HGNC:838] | 30323 | -0.562 | -0.5788 | Yes |
| 103 | TIMM8B | translocase of inner mitochondrial membrane 8 homolog B [Source:HGNC Symbol;Acc:HGNC:11818] | 30418 | -0.589 | -0.5785 | Yes |
| 104 | VDAC3 | voltage dependent anion channel 3 [Source:HGNC Symbol;Acc:HGNC:12674] | 30433 | -0.596 | -0.5760 | Yes |
| 105 | NDUFS1 | NADH:ubiquinone oxidoreductase core subunit S1 [Source:HGNC Symbol;Acc:HGNC:7707] | 30442 | -0.599 | -0.5733 | Yes |
| 106 | SDHC | succinate dehydrogenase complex subunit C [Source:HGNC Symbol;Acc:HGNC:10682] | 30476 | -0.614 | -0.5712 | Yes |
| 107 | COX5A | cytochrome c oxidase subunit 5A [Source:HGNC Symbol;Acc:HGNC:2267] | 30575 | -0.643 | -0.5707 | Yes |
| 108 | CYB5R3 | cytochrome b5 reductase 3 [Source:HGNC Symbol;Acc:HGNC:2873] | 30588 | -0.647 | -0.5679 | Yes |
| 109 | MTRF1 | mitochondrial translation release factor 1 [Source:HGNC Symbol;Acc:HGNC:7469] | 30620 | -0.657 | -0.5656 | Yes |
| 110 | NDUFA8 | NADH:ubiquinone oxidoreductase subunit A8 [Source:HGNC Symbol;Acc:HGNC:7692] | 30650 | -0.669 | -0.5631 | Yes |
| 111 | PDHX | pyruvate dehydrogenase complex component X [Source:HGNC Symbol;Acc:HGNC:21350] | 30657 | -0.671 | -0.5601 | Yes |
| 112 | ACADVL | acyl-CoA dehydrogenase very long chain [Source:HGNC Symbol;Acc:HGNC:92] | 30784 | -0.705 | -0.5601 | Yes |
| 113 | COX5B | cytochrome c oxidase subunit 5B [Source:HGNC Symbol;Acc:HGNC:2269] | 30804 | -0.711 | -0.5571 | Yes |
| 114 | ABCB7 | ATP binding cassette subfamily B member 7 [Source:HGNC Symbol;Acc:HGNC:48] | 30822 | -0.717 | -0.5541 | Yes |
| 115 | ATP5F1D | ATP synthase F1 subunit delta [Source:HGNC Symbol;Acc:HGNC:837] | 30884 | -0.735 | -0.5522 | Yes |
| 116 | NDUFV1 | NADH:ubiquinone oxidoreductase core subunit V1 [Source:HGNC Symbol;Acc:HGNC:7716] | 30890 | -0.737 | -0.5488 | Yes |
| 117 | GOT2 | glutamic-oxaloacetic transaminase 2 [Source:HGNC Symbol;Acc:HGNC:4433] | 30938 | -0.749 | -0.5464 | Yes |
| 118 | ISCU | iron-sulfur cluster assembly enzyme [Source:HGNC Symbol;Acc:HGNC:29882] | 30996 | -0.769 | -0.5443 | Yes |
| 119 | NDUFS8 | NADH:ubiquinone oxidoreductase core subunit S8 [Source:HGNC Symbol;Acc:HGNC:7715] | 30997 | -0.769 | -0.5405 | Yes |
| 120 | UQCRC1 | ubiquinol-cytochrome c reductase core protein 1 [Source:HGNC Symbol;Acc:HGNC:12585] | 31029 | -0.781 | -0.5376 | Yes |
| 121 | UQCRFS1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 [Source:HGNC Symbol;Acc:HGNC:12587] | 31146 | -0.807 | -0.5368 | Yes |
| 122 | NDUFA7 | NADH:ubiquinone oxidoreductase subunit A7 [Source:HGNC Symbol;Acc:HGNC:7691] | 31184 | -0.820 | -0.5339 | Yes |
| 123 | MAOB | monoamine oxidase B [Source:HGNC Symbol;Acc:HGNC:6834] | 31195 | -0.825 | -0.5302 | Yes |
| 124 | ATP6AP1 | ATPase H+ transporting accessory protein 1 [Source:HGNC Symbol;Acc:HGNC:868] | 31206 | -0.830 | -0.5264 | Yes |
| 125 | MRPS30 | mitochondrial ribosomal protein S30 [Source:HGNC Symbol;Acc:HGNC:8769] | 31242 | -0.842 | -0.5233 | Yes |
| 126 | RHOT2 | ras homolog family member T2 [Source:HGNC Symbol;Acc:HGNC:21169] | 31287 | -0.857 | -0.5203 | Yes |
| 127 | AFG3L2 | AFG3 like matrix AAA peptidase subunit 2 [Source:HGNC Symbol;Acc:HGNC:315] | 31389 | -0.889 | -0.5188 | Yes |
| 128 | NDUFC2 | NADH:ubiquinone oxidoreductase subunit C2 [Source:HGNC Symbol;Acc:HGNC:7706] | 31446 | -0.909 | -0.5159 | Yes |
| 129 | MPC1 | mitochondrial pyruvate carrier 1 [Source:HGNC Symbol;Acc:HGNC:21606] | 31583 | -0.949 | -0.5150 | Yes |
| 130 | RHOT1 | ras homolog family member T1 [Source:HGNC Symbol;Acc:HGNC:21168] | 31647 | -0.973 | -0.5120 | Yes |
| 131 | BAX | BCL2 associated X, apoptosis regulator [Source:HGNC Symbol;Acc:HGNC:959] | 31717 | -0.999 | -0.5090 | Yes |
| 132 | ATP5MC2 | ATP synthase membrane subunit c locus 2 [Source:HGNC Symbol;Acc:HGNC:842] | 31727 | -1.001 | -0.5044 | Yes |
| 133 | UQCRH | ubiquinol-cytochrome c reductase hinge protein [Source:HGNC Symbol;Acc:HGNC:12590] | 31735 | -1.003 | -0.4998 | Yes |
| 134 | POLR2F | RNA polymerase II subunit F [Source:HGNC Symbol;Acc:HGNC:9193] | 31776 | -1.017 | -0.4959 | Yes |
| 135 | TIMM13 | translocase of inner mitochondrial membrane 13 [Source:HGNC Symbol;Acc:HGNC:11816] | 31825 | -1.032 | -0.4922 | Yes |
| 136 | GLUD1 | glutamate dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:4335] | 31836 | -1.035 | -0.4875 | Yes |
| 137 | CYB5A | cytochrome b5 type A [Source:HGNC Symbol;Acc:HGNC:2570] | 31915 | -1.063 | -0.4845 | Yes |
| 138 | NDUFB7 | NADH:ubiquinone oxidoreductase subunit B7 [Source:HGNC Symbol;Acc:HGNC:7702] | 31933 | -1.069 | -0.4797 | Yes |
| 139 | NDUFA6 | NADH:ubiquinone oxidoreductase subunit A6 [Source:HGNC Symbol;Acc:HGNC:7690] | 31975 | -1.084 | -0.4756 | Yes |
| 140 | PHB2 | prohibitin 2 [Source:HGNC Symbol;Acc:HGNC:30306] | 31979 | -1.085 | -0.4704 | Yes |
| 141 | NDUFS3 | NADH:ubiquinone oxidoreductase core subunit S3 [Source:HGNC Symbol;Acc:HGNC:7710] | 32008 | -1.097 | -0.4659 | Yes |
| 142 | SDHB | succinate dehydrogenase complex iron sulfur subunit B [Source:HGNC Symbol;Acc:HGNC:10681] | 32040 | -1.109 | -0.4614 | Yes |
| 143 | ETFB | electron transfer flavoprotein subunit beta [Source:HGNC Symbol;Acc:HGNC:3482] | 32118 | -1.145 | -0.4579 | Yes |
| 144 | CS | citrate synthase [Source:HGNC Symbol;Acc:HGNC:2422] | 32123 | -1.147 | -0.4525 | Yes |
| 145 | PDHA1 | pyruvate dehydrogenase E1 subunit alpha 1 [Source:HGNC Symbol;Acc:HGNC:8806] | 32168 | -1.165 | -0.4480 | Yes |
| 146 | IDH3B | isocitrate dehydrogenase (NAD(+)) 3 non-catalytic subunit beta [Source:HGNC Symbol;Acc:HGNC:5385] | 32211 | -1.181 | -0.4435 | Yes |
| 147 | SLC25A11 | solute carrier family 25 member 11 [Source:HGNC Symbol;Acc:HGNC:10981] | 32512 | -1.234 | -0.4456 | Yes |
| 148 | MRPS15 | mitochondrial ribosomal protein S15 [Source:HGNC Symbol;Acc:HGNC:14504] | 32613 | -1.274 | -0.4422 | Yes |
| 149 | COX10 | cytochrome c oxidase assembly factor heme A:farnesyltransferase COX10 [Source:HGNC Symbol;Acc:HGNC:2260] | 32648 | -1.291 | -0.4368 | Yes |
| 150 | SUPV3L1 | Suv3 like RNA helicase [Source:HGNC Symbol;Acc:HGNC:11471] | 32690 | -1.315 | -0.4316 | Yes |
| 151 | NDUFS2 | NADH:ubiquinone oxidoreductase core subunit S2 [Source:HGNC Symbol;Acc:HGNC:7708] | 32699 | -1.320 | -0.4254 | Yes |
| 152 | COX7A2L | cytochrome c oxidase subunit 7A2 like [Source:HGNC Symbol;Acc:HGNC:2289] | 32732 | -1.333 | -0.4198 | Yes |
| 153 | OPA1 | OPA1 mitochondrial dynamin like GTPase [Source:HGNC Symbol;Acc:HGNC:8140] | 32738 | -1.336 | -0.4135 | Yes |
| 154 | IDH3A | isocitrate dehydrogenase (NAD(+)) 3 catalytic subunit alpha [Source:HGNC Symbol;Acc:HGNC:5384] | 32794 | -1.356 | -0.4084 | Yes |
| 155 | OGDH | oxoglutarate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:8124] | 32809 | -1.359 | -0.4022 | Yes |
| 156 | IDH3G | isocitrate dehydrogenase (NAD(+)) 3 non-catalytic subunit gamma [Source:HGNC Symbol;Acc:HGNC:5386] | 32912 | -1.405 | -0.3982 | Yes |
| 157 | ETFDH | electron transfer flavoprotein dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3483] | 33158 | -1.512 | -0.3975 | Yes |
| 158 | SDHA | succinate dehydrogenase complex flavoprotein subunit A [Source:HGNC Symbol;Acc:HGNC:10680] | 33296 | -1.574 | -0.3936 | Yes |
| 159 | PHYH | phytanoyl-CoA 2-hydroxylase [Source:HGNC Symbol;Acc:HGNC:8940] | 33345 | -1.592 | -0.3872 | Yes |
| 160 | FH | fumarate hydratase [Source:HGNC Symbol;Acc:HGNC:3700] | 33386 | -1.614 | -0.3805 | Yes |
| 161 | ATP5F1C | ATP synthase F1 subunit gamma [Source:HGNC Symbol;Acc:HGNC:833] | 33391 | -1.618 | -0.3728 | Yes |
| 162 | SLC25A4 | solute carrier family 25 member 4 [Source:HGNC Symbol;Acc:HGNC:10990] | 33424 | -1.630 | -0.3657 | Yes |
| 163 | NDUFS7 | NADH:ubiquinone oxidoreductase core subunit S7 [Source:HGNC Symbol;Acc:HGNC:7714] | 33437 | -1.635 | -0.3582 | Yes |
| 164 | NDUFV2 | NADH:ubiquinone oxidoreductase core subunit V2 [Source:HGNC Symbol;Acc:HGNC:7717] | 33455 | -1.646 | -0.3506 | Yes |
| 165 | NDUFB8 | NADH:ubiquinone oxidoreductase subunit B8 [Source:HGNC Symbol;Acc:HGNC:7703] | 33469 | -1.650 | -0.3430 | Yes |
| 166 | HADHB | hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta [Source:HGNC Symbol;Acc:HGNC:4803] | 33471 | -1.650 | -0.3351 | Yes |
| 167 | LRPPRC | leucine rich pentatricopeptide repeat containing [Source:HGNC Symbol;Acc:HGNC:15714] | 33527 | -1.679 | -0.3284 | Yes |
| 168 | ACAT1 | acetyl-CoA acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:93] | 33726 | -1.764 | -0.3252 | Yes |
| 169 | MDH1 | malate dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:6970] | 33776 | -1.797 | -0.3179 | Yes |
| 170 | NQO2 | N-ribosyldihydronicotinamide:quinone reductase 2 [Source:HGNC Symbol;Acc:HGNC:7856] | 33878 | -1.854 | -0.3116 | Yes |
| 171 | ECI1 | enoyl-CoA delta isomerase 1 [Source:HGNC Symbol;Acc:HGNC:2703] | 33932 | -1.880 | -0.3040 | Yes |
| 172 | SLC25A6 | solute carrier family 25 member 6 [Source:HGNC Symbol;Acc:HGNC:10992] | 33950 | -1.884 | -0.2953 | Yes |
| 173 | OAT | ornithine aminotransferase [Source:HGNC Symbol;Acc:HGNC:8091] | 34191 | -2.020 | -0.2921 | Yes |
| 174 | LDHA | lactate dehydrogenase A [Source:HGNC Symbol;Acc:HGNC:6535] | 34477 | -2.137 | -0.2894 | Yes |
| 175 | ACADM | acyl-CoA dehydrogenase medium chain [Source:HGNC Symbol;Acc:HGNC:89] | 34543 | -2.191 | -0.2806 | Yes |
| 176 | FXN | frataxin [Source:HGNC Symbol;Acc:HGNC:3951] | 34605 | -2.237 | -0.2714 | Yes |
| 177 | MRPL34 | mitochondrial ribosomal protein L34 [Source:HGNC Symbol;Acc:HGNC:14488] | 34699 | -2.288 | -0.2629 | Yes |
| 178 | SURF1 | SURF1 cytochrome c oxidase assembly factor [Source:HGNC Symbol;Acc:HGNC:11474] | 34709 | -2.294 | -0.2520 | Yes |
| 179 | OXA1L | OXA1L mitochondrial inner membrane protein [Source:HGNC Symbol;Acc:HGNC:8526] | 34760 | -2.320 | -0.2421 | Yes |
| 180 | ECHS1 | enoyl-CoA hydratase, short chain 1 [Source:HGNC Symbol;Acc:HGNC:3151] | 34764 | -2.324 | -0.2310 | Yes |
| 181 | BCKDHA | branched chain keto acid dehydrogenase E1 subunit alpha [Source:HGNC Symbol;Acc:HGNC:986] | 34830 | -2.376 | -0.2212 | Yes |
| 182 | ALDH6A1 | aldehyde dehydrogenase 6 family member A1 [Source:HGNC Symbol;Acc:HGNC:7179] | 34878 | -2.406 | -0.2109 | Yes |
| 183 | ACAA1 | acetyl-CoA acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:82] | 34902 | -2.428 | -0.1997 | Yes |
| 184 | GPI | glucose-6-phosphate isomerase [Source:HGNC Symbol;Acc:HGNC:4458] | 35013 | -2.494 | -0.1907 | Yes |
| 185 | HSD17B10 | hydroxysteroid 17-beta dehydrogenase 10 [Source:HGNC Symbol;Acc:HGNC:4800] | 35050 | -2.525 | -0.1794 | Yes |
| 186 | RETSAT | retinol saturase [Source:HGNC Symbol;Acc:HGNC:25991] | 35106 | -2.563 | -0.1685 | Yes |
| 187 | IDH2 | isocitrate dehydrogenase (NADP(+)) 2 [Source:HGNC Symbol;Acc:HGNC:5383] | 35196 | -2.643 | -0.1581 | Yes |
| 188 | FDX1 | ferredoxin 1 [Source:HGNC Symbol;Acc:HGNC:3638] | 35250 | -2.679 | -0.1466 | Yes |
| 189 | CPT1A | carnitine palmitoyltransferase 1A [Source:HGNC Symbol;Acc:HGNC:2328] | 35438 | -2.788 | -0.1382 | Yes |
| 190 | VDAC1 | voltage dependent anion channel 1 [Source:HGNC Symbol;Acc:HGNC:12669] | 35500 | -2.844 | -0.1261 | Yes |
| 191 | MGST3 | microsomal glutathione S-transferase 3 [Source:HGNC Symbol;Acc:HGNC:7064] | 35519 | -2.863 | -0.1127 | Yes |
| 192 | HTRA2 | HtrA serine peptidase 2 [Source:HGNC Symbol;Acc:HGNC:14348] | 35559 | -2.897 | -0.0997 | Yes |
| 193 | NNT | nicotinamide nucleotide transhydrogenase [Source:HGNC Symbol;Acc:HGNC:7863] | 35812 | -3.155 | -0.0913 | Yes |
| 194 | NDUFC1 | NADH:ubiquinone oxidoreductase subunit C1 [Source:HGNC Symbol;Acc:HGNC:7705] | 36279 | -3.606 | -0.0865 | Yes |
| 195 | GPX4 | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 36315 | -3.646 | -0.0698 | Yes |
| 196 | ECH1 | enoyl-CoA hydratase 1 [Source:HGNC Symbol;Acc:HGNC:3149] | 36319 | -3.655 | -0.0522 | Yes |
| 197 | ACAA2 | acetyl-CoA acyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:83] | 36335 | -3.680 | -0.0348 | Yes |
| 198 | ATP5MF | ATP synthase membrane subunit f [Source:HGNC Symbol;Acc:HGNC:848] | 36411 | -3.773 | -0.0185 | Yes |
| 199 | PDP1 | pyruvate dehyrogenase phosphatase catalytic subunit 1 [Source:HGNC Symbol;Acc:HGNC:9279] | 36418 | -3.780 | -0.0004 | Yes |
| 200 | IDH1 | isocitrate dehydrogenase (NADP(+)) 1 [Source:HGNC Symbol;Acc:HGNC:5382] | 36479 | -3.842 | 0.0166 | Yes |