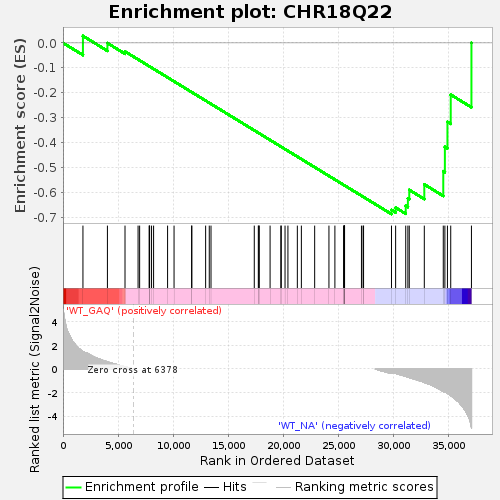
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #WT_GAQ_versus_WT_NA.Nof1.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | Nof1.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
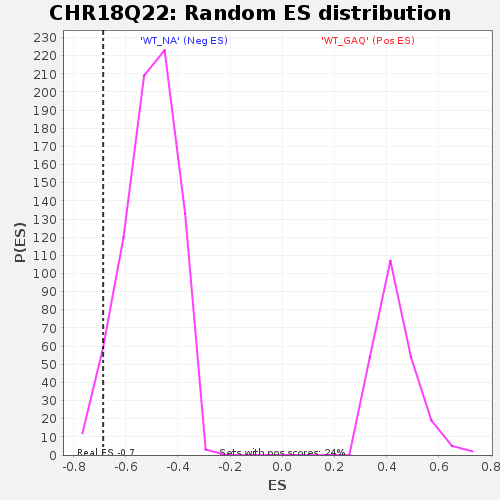
| GeneSet | CHR18Q22 |
| Enrichment Score (ES) | -0.68649983 |
| Normalized Enrichment Score (NES) | -1.3530549 |
| Nominal p-value | 0.04611331 |
| FDR q-value | 0.37902483 |
| FWER p-Value | 0.995 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CNDP2 | carnosine dipeptidase 2 [Source:HGNC Symbol;Acc:HGNC:24437] | 1810 | 1.472 | 0.0280 | No |
| 2 | SOCS6 | suppressor of cytokine signaling 6 [Source:HGNC Symbol;Acc:HGNC:16833] | 4032 | 0.597 | -0.0008 | No |
| 3 | ZNF407 | zinc finger protein 407 [Source:HGNC Symbol;Acc:HGNC:19904] | 5629 | 0.182 | -0.0344 | No |
| 4 | MIR548AV | microRNA 548av [Source:HGNC Symbol;Acc:HGNC:43537] | 6815 | 0.000 | -0.0664 | No |
| 5 | DIPK1C | divergent protein kinase domain 1C [Source:HGNC Symbol;Acc:HGNC:31729] | 6957 | 0.000 | -0.0702 | No |
| 6 | RPL12P39 | ribosomal protein L12 pseudogene 39 [Source:HGNC Symbol;Acc:HGNC:35976] | 7825 | 0.000 | -0.0936 | No |
| 7 | LINC01541 | long intergenic non-protein coding RNA 1541 [Source:HGNC Symbol;Acc:HGNC:51309] | 7843 | 0.000 | -0.0941 | No |
| 8 | LINC01538 | long intergenic non-protein coding RNA 1538 [Source:HGNC Symbol;Acc:HGNC:51306] | 7863 | 0.000 | -0.0946 | No |
| 9 | NETO1 | neuropilin and tolloid like 1 [Source:HGNC Symbol;Acc:HGNC:13823] | 8038 | 0.000 | -0.0993 | No |
| 10 | CD226 | CD226 molecule [Source:HGNC Symbol;Acc:HGNC:16961] | 8224 | 0.000 | -0.1043 | No |
| 11 | CDH7 | cadherin 7 [Source:HGNC Symbol;Acc:HGNC:1766] | 9494 | 0.000 | -0.1386 | No |
| 12 | AKR1B10P2 | aldo-keto reductase family 1 member B10 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:45063] | 10081 | 0.000 | -0.1544 | No |
| 13 | RN7SL401P | RNA, 7SL, cytoplasmic 401, pseudogene [Source:HGNC Symbol;Acc:HGNC:46417] | 11673 | 0.000 | -0.1973 | No |
| 14 | MIR5011 | microRNA 5011 [Source:HGNC Symbol;Acc:HGNC:43496] | 11698 | 0.000 | -0.1980 | No |
| 15 | LINC00305 | long intergenic non-protein coding RNA 305 [Source:HGNC Symbol;Acc:HGNC:28597] | 12948 | 0.000 | -0.2317 | No |
| 16 | AC119868.1 | cholinergic receptor, nicotinic, epsilon (CHRNE) pseudogene | 13278 | 0.000 | -0.2406 | No |
| 17 | CBLN2 | cerebellin 2 precursor [Source:HGNC Symbol;Acc:HGNC:1544] | 13436 | 0.000 | -0.2448 | No |
| 18 | RNA5SP460 | RNA, 5S ribosomal pseudogene 460 [Source:HGNC Symbol;Acc:HGNC:43360] | 17364 | 0.000 | -0.3509 | No |
| 19 | DOK6 | docking protein 6 [Source:HGNC Symbol;Acc:HGNC:28301] | 17731 | 0.000 | -0.3607 | No |
| 20 | RN7SL795P | RNA, 7SL, cytoplasmic 795, pseudogene [Source:HGNC Symbol;Acc:HGNC:46811] | 17787 | 0.000 | -0.3622 | No |
| 21 | RN7SL551P | RNA, 7SL, cytoplasmic 551, pseudogene [Source:HGNC Symbol;Acc:HGNC:46567] | 17807 | 0.000 | -0.3627 | No |
| 22 | CDH19 | cadherin 19 [Source:HGNC Symbol;Acc:HGNC:1758] | 18799 | 0.000 | -0.3895 | No |
| 23 | LINC01899 | long intergenic non-protein coding RNA 1899 [Source:HGNC Symbol;Acc:HGNC:52718] | 19775 | 0.000 | -0.4158 | No |
| 24 | AC091138.1 | novel transcript | 19815 | 0.000 | -0.4169 | No |
| 25 | LINC00909 | long intergenic non-protein coding RNA 909 [Source:HGNC Symbol;Acc:HGNC:44331] | 20156 | 0.000 | -0.4260 | No |
| 26 | CNDP1 | carnosine dipeptidase 1 [Source:HGNC Symbol;Acc:HGNC:20675] | 20419 | 0.000 | -0.4331 | No |
| 27 | PRPF19P1 | PRPF19 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:23182] | 21273 | 0.000 | -0.4561 | No |
| 28 | C18orf63 | chromosome 18 open reading frame 63 [Source:HGNC Symbol;Acc:HGNC:40037] | 21639 | 0.000 | -0.4660 | No |
| 29 | HMSD | histocompatibility minor serpin domain containing [Source:HGNC Symbol;Acc:HGNC:23037] | 22846 | 0.000 | -0.4986 | No |
| 30 | GTSCR1 | Gilles de la Tourette syndrome chromosome region, candidate 1 [Source:HGNC Symbol;Acc:HGNC:18406] | 24142 | 0.000 | -0.5335 | No |
| 31 | RNU6-1037P | RNA, U6 small nuclear 1037, pseudogene [Source:HGNC Symbol;Acc:HGNC:48000] | 24682 | 0.000 | -0.5481 | No |
| 32 | LINC01909 | long intergenic non-protein coding RNA 1909 [Source:HGNC Symbol;Acc:HGNC:52728] | 25488 | 0.000 | -0.5698 | No |
| 33 | LINC01903 | long intergenic non-protein coding RNA 1903 [Source:HGNC Symbol;Acc:HGNC:52722] | 25491 | 0.000 | -0.5699 | No |
| 34 | LINC01924 | long intergenic non-protein coding RNA 1924 [Source:HGNC Symbol;Acc:HGNC:27600] | 25529 | 0.000 | -0.5709 | No |
| 35 | LINC01922 | long intergenic non-protein coding RNA 1922 [Source:HGNC Symbol;Acc:HGNC:52741] | 25539 | 0.000 | -0.5711 | No |
| 36 | LINC01916 | long intergenic non-protein coding RNA 1916 [Source:HGNC Symbol;Acc:HGNC:52735] | 25554 | 0.000 | -0.5715 | No |
| 37 | LINC01910 | long intergenic non-protein coding RNA 1910 [Source:HGNC Symbol;Acc:HGNC:52729] | 25557 | 0.000 | -0.5715 | No |
| 38 | LINC01912 | long intergenic non-protein coding RNA 1912 [Source:HGNC Symbol;Acc:HGNC:52731] | 25558 | 0.000 | -0.5715 | No |
| 39 | LIVAR | liver cell viability associated lncRNA [Source:HGNC Symbol;Acc:HGNC:53130] | 27093 | 0.000 | -0.6130 | No |
| 40 | CCDC102B | coiled-coil domain containing 102B [Source:HGNC Symbol;Acc:HGNC:26295] | 27240 | 0.000 | -0.6169 | No |
| 41 | LARP7P3 | LARP7 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:49765] | 27285 | 0.000 | -0.6181 | No |
| 42 | TIMM21 | translocase of inner mitochondrial membrane 21 [Source:HGNC Symbol;Acc:HGNC:25010] | 29820 | -0.320 | -0.6698 | Yes |
| 43 | TMX3 | thioredoxin related transmembrane protein 3 [Source:HGNC Symbol;Acc:HGNC:24718] | 30196 | -0.371 | -0.6605 | Yes |
| 44 | ZADH2 | zinc binding alcohol dehydrogenase domain containing 2 [Source:HGNC Symbol;Acc:HGNC:28697] | 31113 | -0.618 | -0.6530 | Yes |
| 45 | TSHZ1 | teashirt zinc finger homeobox 1 [Source:HGNC Symbol;Acc:HGNC:10669] | 31309 | -0.672 | -0.6232 | Yes |
| 46 | RTTN | rotatin [Source:HGNC Symbol;Acc:HGNC:18654] | 31434 | -0.708 | -0.5895 | Yes |
| 47 | CYB5A | cytochrome b5 type A [Source:HGNC Symbol;Acc:HGNC:2570] | 32799 | -1.130 | -0.5674 | Yes |
| 48 | SERPINB8 | serpin family B member 8 [Source:HGNC Symbol;Acc:HGNC:8952] | 34521 | -1.903 | -0.5145 | Yes |
| 49 | FBXO15 | F-box protein 15 [Source:HGNC Symbol;Acc:HGNC:13617] | 34667 | -1.945 | -0.4169 | Yes |
| 50 | HNRNPA1P11 | heterogeneous nuclear ribonucleoprotein A1 pseudogene 11 [Source:HGNC Symbol;Acc:HGNC:39129] | 34899 | -2.039 | -0.3167 | Yes |
| 51 | DSEL | dermatan sulfate epimerase like [Source:HGNC Symbol;Acc:HGNC:18144] | 35198 | -2.243 | -0.2076 | Yes |
| 52 | RNU6-39P | RNA, U6 small nuclear 39, pseudogene [Source:HGNC Symbol;Acc:HGNC:34283] | 37078 | -4.955 | 0.0003 | Yes |