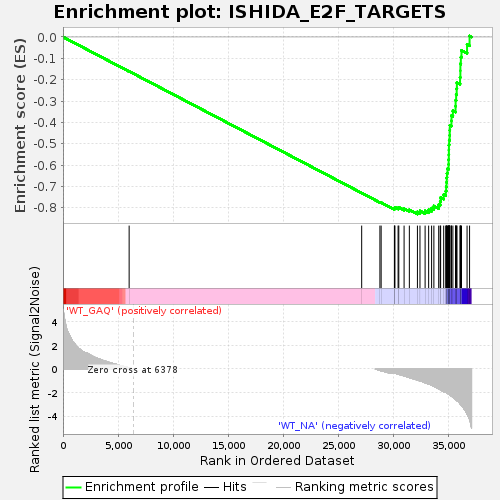
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #WT_GAQ_versus_WT_NA.Nof1.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | Nof1.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | ISHIDA_E2F_TARGETS |
| Enrichment Score (ES) | -0.82968825 |
| Normalized Enrichment Score (NES) | -1.6386971 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.028282123 |
| FWER p-Value | 0.347 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | DCTPP1 | dCTP pyrophosphatase 1 [Source:HGNC Symbol;Acc:HGNC:28777] | 6004 | 0.097 | -0.1610 | No |
| 2 | ANXA8L1 | annexin A8 like 1 [Source:HGNC Symbol;Acc:HGNC:23334] | 27114 | 0.000 | -0.7309 | No |
| 3 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 28769 | -0.115 | -0.7743 | No |
| 4 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 28881 | -0.144 | -0.7758 | No |
| 5 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 30083 | -0.340 | -0.8045 | No |
| 6 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 30110 | -0.349 | -0.8014 | No |
| 7 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 30121 | -0.351 | -0.7979 | No |
| 8 | CCNB1 | cyclin B1 [Source:HGNC Symbol;Acc:HGNC:1579] | 30420 | -0.426 | -0.8013 | No |
| 9 | KPNA2 | karyopherin subunit alpha 2 [Source:HGNC Symbol;Acc:HGNC:6395] | 30483 | -0.442 | -0.7981 | No |
| 10 | CDT1 | chromatin licensing and DNA replication factor 1 [Source:HGNC Symbol;Acc:HGNC:24576] | 30964 | -0.580 | -0.8048 | No |
| 11 | SLBP | stem-loop binding protein [Source:HGNC Symbol;Acc:HGNC:10904] | 31441 | -0.711 | -0.8099 | No |
| 12 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 32174 | -0.931 | -0.8196 | Yes |
| 13 | ASF1B | anti-silencing function 1B histone chaperone [Source:HGNC Symbol;Acc:HGNC:20996] | 32405 | -1.003 | -0.8149 | Yes |
| 14 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 32872 | -1.155 | -0.8149 | Yes |
| 15 | RAD51 | RAD51 recombinase [Source:HGNC Symbol;Acc:HGNC:9817] | 33193 | -1.285 | -0.8096 | Yes |
| 16 | CDCA7 | cell division cycle associated 7 [Source:HGNC Symbol;Acc:HGNC:14628] | 33461 | -1.370 | -0.8019 | Yes |
| 17 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 33668 | -1.457 | -0.7916 | Yes |
| 18 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 34106 | -1.671 | -0.7852 | Yes |
| 19 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 34241 | -1.737 | -0.7699 | Yes |
| 20 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 34281 | -1.756 | -0.7519 | Yes |
| 21 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit [Source:HGNC Symbol;Acc:HGNC:3527] | 34559 | -1.929 | -0.7384 | Yes |
| 22 | CDK2 | cyclin dependent kinase 2 [Source:HGNC Symbol;Acc:HGNC:1771] | 34747 | -1.948 | -0.7223 | Yes |
| 23 | LBR | lamin B receptor [Source:HGNC Symbol;Acc:HGNC:6518] | 34785 | -1.968 | -0.7019 | Yes |
| 24 | DNA2 | DNA replication helicase/nuclease 2 [Source:HGNC Symbol;Acc:HGNC:2939] | 34818 | -1.991 | -0.6811 | Yes |
| 25 | UBE2T | ubiquitin conjugating enzyme E2 T [Source:HGNC Symbol;Acc:HGNC:25009] | 34835 | -2.003 | -0.6597 | Yes |
| 26 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 34857 | -2.014 | -0.6384 | Yes |
| 27 | ANLN | anillin actin binding protein [Source:HGNC Symbol;Acc:HGNC:14082] | 34904 | -2.042 | -0.6174 | Yes |
| 28 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 35003 | -2.106 | -0.5972 | Yes |
| 29 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35009 | -2.109 | -0.5744 | Yes |
| 30 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35024 | -2.115 | -0.5518 | Yes |
| 31 | DBF4 | DBF4 zinc finger [Source:HGNC Symbol;Acc:HGNC:17364] | 35030 | -2.121 | -0.5288 | Yes |
| 32 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 35034 | -2.123 | -0.5058 | Yes |
| 33 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 35057 | -2.142 | -0.4831 | Yes |
| 34 | SGO1 | shugoshin 1 [Source:HGNC Symbol;Acc:HGNC:25088] | 35101 | -2.164 | -0.4608 | Yes |
| 35 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35109 | -2.175 | -0.4373 | Yes |
| 36 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 35118 | -2.180 | -0.4138 | Yes |
| 37 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 35261 | -2.280 | -0.3928 | Yes |
| 38 | BUB1 | BUB1 mitotic checkpoint serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1148] | 35269 | -2.283 | -0.3682 | Yes |
| 39 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 35394 | -2.370 | -0.3458 | Yes |
| 40 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 35634 | -2.552 | -0.3245 | Yes |
| 41 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 35642 | -2.564 | -0.2968 | Yes |
| 42 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 35689 | -2.604 | -0.2697 | Yes |
| 43 | CCNE1 | cyclin E1 [Source:HGNC Symbol;Acc:HGNC:1589] | 35738 | -2.643 | -0.2422 | Yes |
| 44 | HMGB3 | high mobility group box 3 [Source:HGNC Symbol;Acc:HGNC:5004] | 35757 | -2.659 | -0.2138 | Yes |
| 45 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36044 | -2.930 | -0.1897 | Yes |
| 46 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36073 | -2.962 | -0.1582 | Yes |
| 47 | RPA1 | replication protein A1 [Source:HGNC Symbol;Acc:HGNC:10289] | 36075 | -2.964 | -0.1260 | Yes |
| 48 | CDKN2C | cyclin dependent kinase inhibitor 2C [Source:HGNC Symbol;Acc:HGNC:1789] | 36116 | -2.992 | -0.0946 | Yes |
| 49 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36175 | -3.068 | -0.0628 | Yes |
| 50 | RB1 | RB transcriptional corepressor 1 [Source:HGNC Symbol;Acc:HGNC:9884] | 36681 | -3.791 | -0.0352 | Yes |
| 51 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 36908 | -4.248 | 0.0049 | Yes |

