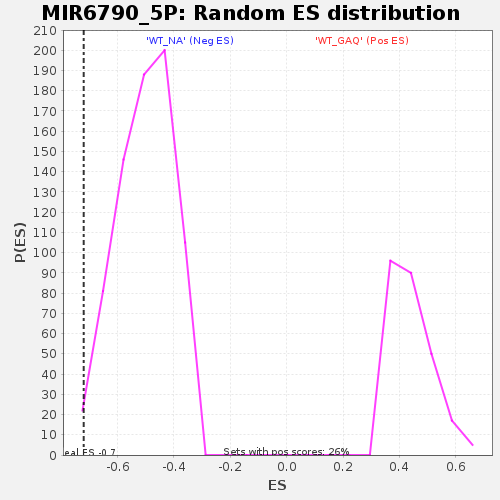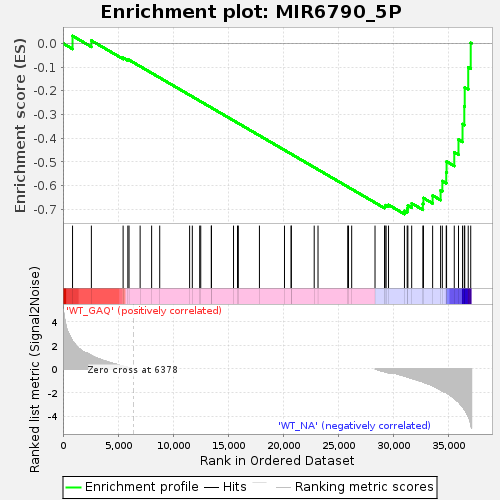
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #WT_GAQ_versus_WT_NA.Nof1.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | Nof1.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | MIR6790_5P |
| Enrichment Score (ES) | -0.72031677 |
| Normalized Enrichment Score (NES) | -1.43544 |
| Nominal p-value | 0.008086253 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MALT1 | MALT1 paracaspase [Source:HGNC Symbol;Acc:HGNC:6819] | 866 | 2.342 | 0.0316 | No |
| 2 | LONRF1 | LON peptidase N-terminal domain and ring finger 1 [Source:HGNC Symbol;Acc:HGNC:26302] | 2576 | 1.127 | 0.0119 | No |
| 3 | GDAP2 | ganglioside induced differentiation associated protein 2 [Source:HGNC Symbol;Acc:HGNC:18010] | 5457 | 0.227 | -0.0605 | No |
| 4 | COL15A1 | collagen type XV alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:2192] | 5887 | 0.126 | -0.0692 | No |
| 5 | ZCCHC2 | zinc finger CCHC-type containing 2 [Source:HGNC Symbol;Acc:HGNC:22916] | 5994 | 0.099 | -0.0697 | No |
| 6 | PRKCQ | protein kinase C theta [Source:HGNC Symbol;Acc:HGNC:9410] | 7002 | 0.000 | -0.0969 | No |
| 7 | EHHADH | enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3247] | 8045 | 0.000 | -0.1250 | No |
| 8 | RASSF9 | Ras association domain family member 9 [Source:HGNC Symbol;Acc:HGNC:15739] | 8780 | 0.000 | -0.1448 | No |
| 9 | LYPLAL1 | lysophospholipase like 1 [Source:HGNC Symbol;Acc:HGNC:20440] | 11494 | 0.000 | -0.2181 | No |
| 10 | PTHLH | parathyroid hormone like hormone [Source:HGNC Symbol;Acc:HGNC:9607] | 11740 | 0.000 | -0.2247 | No |
| 11 | CNMD | chondromodulin [Source:HGNC Symbol;Acc:HGNC:17005] | 12410 | 0.000 | -0.2428 | No |
| 12 | APOBEC3D | apolipoprotein B mRNA editing enzyme catalytic subunit 3D [Source:HGNC Symbol;Acc:HGNC:17354] | 12504 | 0.000 | -0.2453 | No |
| 13 | PDE1A | phosphodiesterase 1A [Source:HGNC Symbol;Acc:HGNC:8774] | 13461 | 0.000 | -0.2711 | No |
| 14 | GDF6 | growth differentiation factor 6 [Source:HGNC Symbol;Acc:HGNC:4221] | 13480 | 0.000 | -0.2716 | No |
| 15 | APELA | apelin receptor early endogenous ligand [Source:HGNC Symbol;Acc:HGNC:48925] | 15480 | 0.000 | -0.3255 | No |
| 16 | CPM | carboxypeptidase M [Source:HGNC Symbol;Acc:HGNC:2311] | 15842 | 0.000 | -0.3353 | No |
| 17 | SLC35F4 | solute carrier family 35 member F4 [Source:HGNC Symbol;Acc:HGNC:19845] | 15915 | 0.000 | -0.3372 | No |
| 18 | SERPINA5 | serpin family A member 5 [Source:HGNC Symbol;Acc:HGNC:8723] | 17832 | 0.000 | -0.3889 | No |
| 19 | INS-IGF2 | INS-IGF2 readthrough [Source:HGNC Symbol;Acc:HGNC:33527] | 20106 | 0.000 | -0.4503 | No |
| 20 | FOXI2 | forkhead box I2 [Source:HGNC Symbol;Acc:HGNC:32448] | 20705 | 0.000 | -0.4664 | No |
| 21 | RIMKLA | ribosomal modification protein rimK like family member A [Source:HGNC Symbol;Acc:HGNC:28725] | 20728 | 0.000 | -0.4670 | No |
| 22 | NLGN4Y | neuroligin 4 Y-linked [Source:HGNC Symbol;Acc:HGNC:15529] | 22804 | 0.000 | -0.5231 | No |
| 23 | STK39 | serine/threonine kinase 39 [Source:HGNC Symbol;Acc:HGNC:17717] | 23150 | 0.000 | -0.5324 | No |
| 24 | ARPP21 | cAMP regulated phosphoprotein 21 [Source:HGNC Symbol;Acc:HGNC:16968] | 25848 | 0.000 | -0.6052 | No |
| 25 | ZNF772 | zinc finger protein 772 [Source:HGNC Symbol;Acc:HGNC:33106] | 25916 | 0.000 | -0.6070 | No |
| 26 | ERICH3 | glutamate rich 3 [Source:HGNC Symbol;Acc:HGNC:25346] | 26206 | 0.000 | -0.6148 | No |
| 27 | SLC2A2 | solute carrier family 2 member 2 [Source:HGNC Symbol;Acc:HGNC:11006] | 28325 | 0.000 | -0.6720 | No |
| 28 | GOLGA1 | golgin A1 [Source:HGNC Symbol;Acc:HGNC:4424] | 29188 | -0.218 | -0.6901 | No |
| 29 | PHF20L1 | PHD finger protein 20 like 1 [Source:HGNC Symbol;Acc:HGNC:24280] | 29233 | -0.228 | -0.6860 | No |
| 30 | TASOR2 | transcription activation suppressor family member 2 [Source:HGNC Symbol;Acc:HGNC:23484] | 29333 | -0.251 | -0.6827 | No |
| 31 | KLHL11 | kelch like family member 11 [Source:HGNC Symbol;Acc:HGNC:19008] | 29551 | -0.311 | -0.6813 | No |
| 32 | PRELID1 | PRELI domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30255] | 30997 | -0.589 | -0.7065 | Yes |
| 33 | SOAT1 | sterol O-acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:11177] | 31246 | -0.652 | -0.6979 | Yes |
| 34 | UBA3 | ubiquitin like modifier activating enzyme 3 [Source:HGNC Symbol;Acc:HGNC:12470] | 31307 | -0.672 | -0.6838 | Yes |
| 35 | WLS | Wnt ligand secretion mediator [Source:HGNC Symbol;Acc:HGNC:30238] | 31653 | -0.775 | -0.6749 | Yes |
| 36 | TCF20 | transcription factor 20 [Source:HGNC Symbol;Acc:HGNC:11631] | 32668 | -1.077 | -0.6770 | Yes |
| 37 | LRPPRC | leucine rich pentatricopeptide repeat containing [Source:HGNC Symbol;Acc:HGNC:15714] | 32722 | -1.102 | -0.6526 | Yes |
| 38 | ARID5B | AT-rich interaction domain 5B [Source:HGNC Symbol;Acc:HGNC:17362] | 33555 | -1.415 | -0.6418 | Yes |
| 39 | RSRP1 | arginine and serine rich protein 1 [Source:HGNC Symbol;Acc:HGNC:25234] | 34278 | -1.755 | -0.6201 | Yes |
| 40 | KBTBD6 | kelch repeat and BTB domain containing 6 [Source:HGNC Symbol;Acc:HGNC:25340] | 34436 | -1.846 | -0.5811 | Yes |
| 41 | KALRN | kalirin RhoGEF kinase [Source:HGNC Symbol;Acc:HGNC:4814] | 34790 | -1.969 | -0.5444 | Yes |
| 42 | OLA1 | Obg like ATPase 1 [Source:HGNC Symbol;Acc:HGNC:28833] | 34821 | -1.995 | -0.4984 | Yes |
| 43 | NSD1 | nuclear receptor binding SET domain protein 1 [Source:HGNC Symbol;Acc:HGNC:14234] | 35519 | -2.460 | -0.4595 | Yes |
| 44 | CALCOCO1 | calcium binding and coiled-coil domain 1 [Source:HGNC Symbol;Acc:HGNC:29306] | 35903 | -2.735 | -0.4057 | Yes |
| 45 | HADH | hydroxyacyl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4799] | 36270 | -3.195 | -0.3406 | Yes |
| 46 | CDC14A | cell division cycle 14A [Source:HGNC Symbol;Acc:HGNC:1718] | 36429 | -3.381 | -0.2655 | Yes |
| 47 | NRN1 | neuritin 1 [Source:HGNC Symbol;Acc:HGNC:17972] | 36469 | -3.443 | -0.1858 | Yes |
| 48 | IDH1 | isocitrate dehydrogenase (NADP(+)) 1 [Source:HGNC Symbol;Acc:HGNC:5382] | 36786 | -3.986 | -0.1008 | Yes |
| 49 | PLXNA4 | plexin A4 [Source:HGNC Symbol;Acc:HGNC:9102] | 37013 | -4.646 | 0.0021 | Yes |