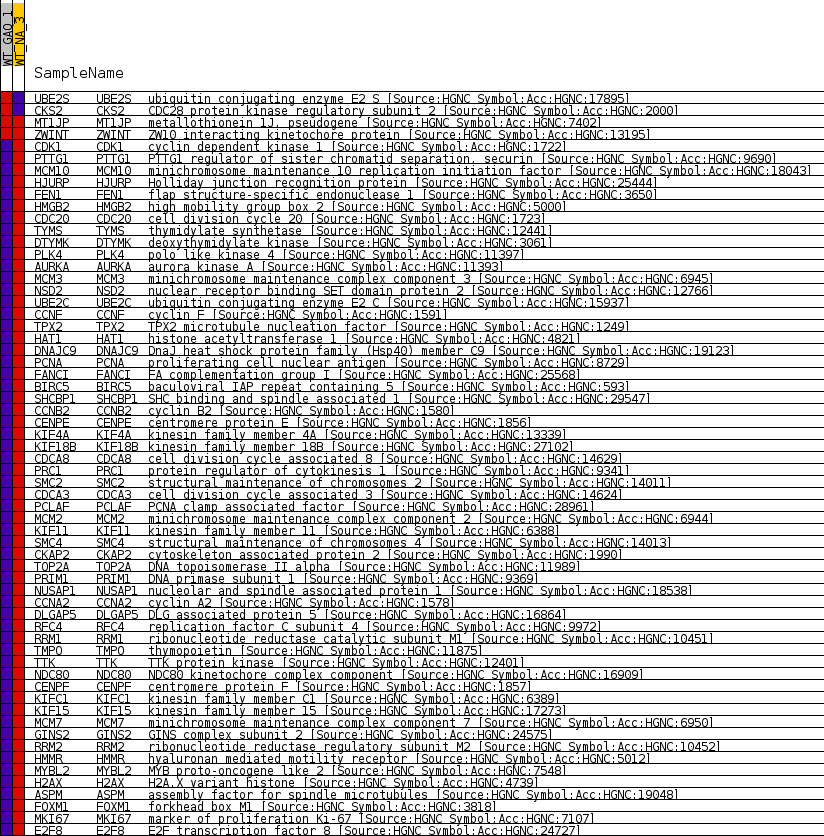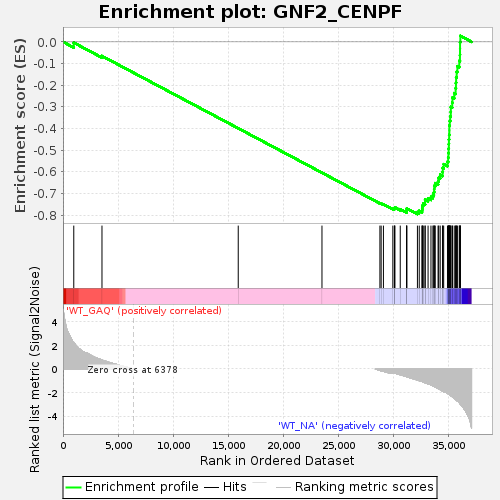
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #WT_GAQ_versus_WT_NA.Nof1.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | Nof1.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | GNF2_CENPF |
| Enrichment Score (ES) | -0.7957044 |
| Normalized Enrichment Score (NES) | -1.6072816 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.016162653 |
| FWER p-Value | 0.152 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | UBE2S | ubiquitin conjugating enzyme E2 S [Source:HGNC Symbol;Acc:HGNC:17895] | 978 | 2.236 | -0.0038 | No |
| 2 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 3538 | 0.752 | -0.0654 | No |
| 3 | MT1JP | metallothionein 1J, pseudogene [Source:HGNC Symbol;Acc:HGNC:7402] | 15908 | 0.000 | -0.3994 | No |
| 4 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 23509 | 0.000 | -0.6046 | No |
| 5 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 28769 | -0.115 | -0.7455 | No |
| 6 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 28881 | -0.144 | -0.7470 | No |
| 7 | MCM10 | minichromosome maintenance 10 replication initiation factor [Source:HGNC Symbol;Acc:HGNC:18043] | 29088 | -0.200 | -0.7506 | No |
| 8 | HJURP | Holliday junction recognition protein [Source:HGNC Symbol;Acc:HGNC:25444] | 29943 | -0.320 | -0.7704 | No |
| 9 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 30083 | -0.340 | -0.7707 | No |
| 10 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 30110 | -0.349 | -0.7679 | No |
| 11 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 30121 | -0.351 | -0.7646 | No |
| 12 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 30615 | -0.478 | -0.7731 | No |
| 13 | DTYMK | deoxythymidylate kinase [Source:HGNC Symbol;Acc:HGNC:3061] | 31195 | -0.639 | -0.7823 | No |
| 14 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 31198 | -0.641 | -0.7759 | No |
| 15 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 31203 | -0.642 | -0.7695 | No |
| 16 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 32174 | -0.931 | -0.7863 | Yes |
| 17 | NSD2 | nuclear receptor binding SET domain protein 2 [Source:HGNC Symbol;Acc:HGNC:12766] | 32339 | -0.980 | -0.7808 | Yes |
| 18 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 32593 | -1.049 | -0.7771 | Yes |
| 19 | CCNF | cyclin F [Source:HGNC Symbol;Acc:HGNC:1591] | 32611 | -1.058 | -0.7669 | Yes |
| 20 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 32628 | -1.064 | -0.7565 | Yes |
| 21 | HAT1 | histone acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:4821] | 32703 | -1.097 | -0.7475 | Yes |
| 22 | DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 [Source:HGNC Symbol;Acc:HGNC:19123] | 32870 | -1.154 | -0.7403 | Yes |
| 23 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 32872 | -1.155 | -0.7287 | Yes |
| 24 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 33141 | -1.259 | -0.7232 | Yes |
| 25 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 33403 | -1.341 | -0.7167 | Yes |
| 26 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 33583 | -1.426 | -0.7071 | Yes |
| 27 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 33668 | -1.457 | -0.6947 | Yes |
| 28 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 33695 | -1.469 | -0.6805 | Yes |
| 29 | KIF4A | kinesin family member 4A [Source:HGNC Symbol;Acc:HGNC:13339] | 33702 | -1.472 | -0.6658 | Yes |
| 30 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 33792 | -1.517 | -0.6529 | Yes |
| 31 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 34050 | -1.644 | -0.6433 | Yes |
| 32 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 34106 | -1.671 | -0.6279 | Yes |
| 33 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 34241 | -1.737 | -0.6140 | Yes |
| 34 | CDCA3 | cell division cycle associated 3 [Source:HGNC Symbol;Acc:HGNC:14624] | 34458 | -1.861 | -0.6010 | Yes |
| 35 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 34472 | -1.873 | -0.5824 | Yes |
| 36 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 34557 | -1.928 | -0.5652 | Yes |
| 37 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 34903 | -2.041 | -0.5539 | Yes |
| 38 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 34967 | -2.085 | -0.5346 | Yes |
| 39 | CKAP2 | cytoskeleton associated protein 2 [Source:HGNC Symbol;Acc:HGNC:1990] | 34988 | -2.096 | -0.5139 | Yes |
| 40 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35009 | -2.109 | -0.4932 | Yes |
| 41 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35024 | -2.115 | -0.4722 | Yes |
| 42 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 35034 | -2.123 | -0.4510 | Yes |
| 43 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 35057 | -2.142 | -0.4300 | Yes |
| 44 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 35073 | -2.150 | -0.4087 | Yes |
| 45 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 35074 | -2.151 | -0.3870 | Yes |
| 46 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35109 | -2.175 | -0.3659 | Yes |
| 47 | TMPO | thymopoietin [Source:HGNC Symbol;Acc:HGNC:11875] | 35150 | -2.203 | -0.3448 | Yes |
| 48 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 35199 | -2.244 | -0.3234 | Yes |
| 49 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 35206 | -2.250 | -0.3008 | Yes |
| 50 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 35334 | -2.329 | -0.2807 | Yes |
| 51 | KIFC1 | kinesin family member C1 [Source:HGNC Symbol;Acc:HGNC:6389] | 35354 | -2.343 | -0.2576 | Yes |
| 52 | KIF15 | kinesin family member 15 [Source:HGNC Symbol;Acc:HGNC:17273] | 35516 | -2.459 | -0.2371 | Yes |
| 53 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 35642 | -2.564 | -0.2146 | Yes |
| 54 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35670 | -2.585 | -0.1892 | Yes |
| 55 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 35689 | -2.604 | -0.1634 | Yes |
| 56 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 35737 | -2.641 | -0.1380 | Yes |
| 57 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 35802 | -2.702 | -0.1125 | Yes |
| 58 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 35972 | -2.829 | -0.0885 | Yes |
| 59 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36035 | -2.919 | -0.0607 | Yes |
| 60 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36042 | -2.928 | -0.0312 | Yes |
| 61 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36044 | -2.930 | -0.0017 | Yes |
| 62 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36073 | -2.962 | 0.0275 | Yes |