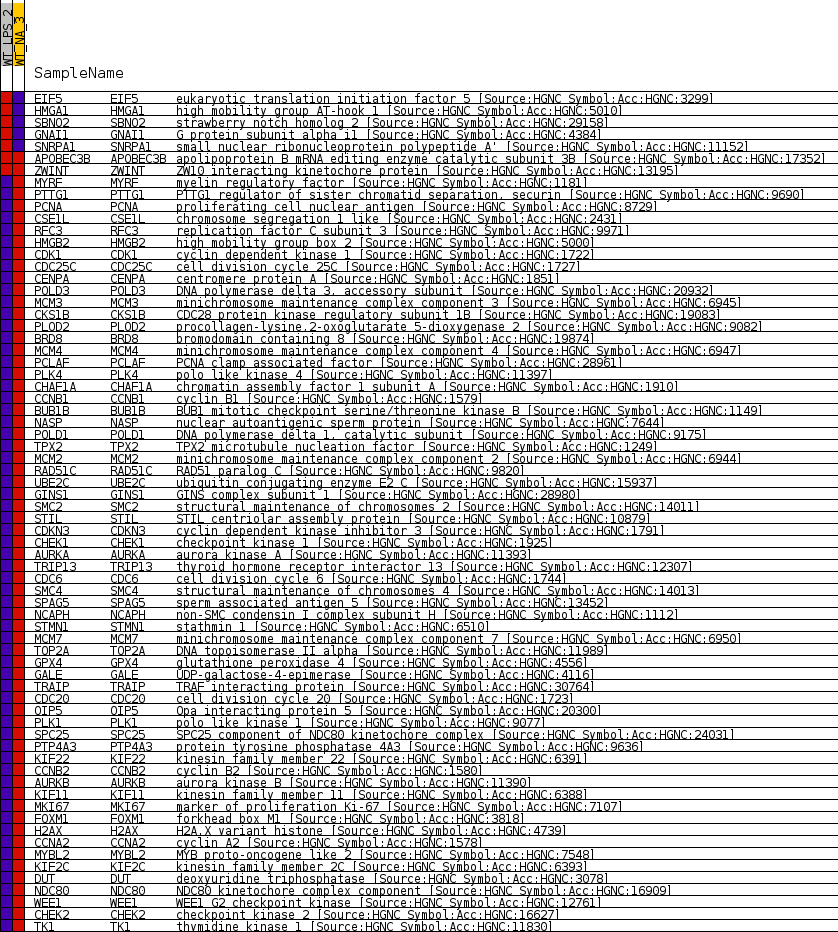
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #WT_LPS_versus_WT_NA.Nof1.cls #WT_LPS_versus_WT_NA_repos |
| Phenotype | Nof1.cls#WT_LPS_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | CROONQUIST_NRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.82991934 |
| Normalized Enrichment Score (NES) | -1.6823348 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.002223426 |
| FWER p-Value | 0.019 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | EIF5 | eukaryotic translation initiation factor 5 [Source:HGNC Symbol;Acc:HGNC:3299] | 3182 | 0.826 | -0.0809 | No |
| 2 | HMGA1 | high mobility group AT-hook 1 [Source:HGNC Symbol;Acc:HGNC:5010] | 3188 | 0.824 | -0.0761 | No |
| 3 | SBNO2 | strawberry notch homolog 2 [Source:HGNC Symbol;Acc:HGNC:29158] | 3346 | 0.761 | -0.0757 | No |
| 4 | GNAI1 | G protein subunit alpha i1 [Source:HGNC Symbol;Acc:HGNC:4384] | 3417 | 0.733 | -0.0731 | No |
| 5 | SNRPA1 | small nuclear ribonucleoprotein polypeptide A' [Source:HGNC Symbol;Acc:HGNC:11152] | 4594 | 0.341 | -0.1028 | No |
| 6 | APOBEC3B | apolipoprotein B mRNA editing enzyme catalytic subunit 3B [Source:HGNC Symbol;Acc:HGNC:17352] | 12359 | 0.000 | -0.3125 | No |
| 7 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 23360 | 0.000 | -0.6096 | No |
| 8 | MYRF | myelin regulatory factor [Source:HGNC Symbol;Acc:HGNC:1181] | 28628 | -0.135 | -0.7511 | No |
| 9 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 30207 | -0.543 | -0.7904 | No |
| 10 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 31440 | -0.872 | -0.8184 | No |
| 11 | CSE1L | chromosome segregation 1 like [Source:HGNC Symbol;Acc:HGNC:2431] | 31492 | -0.894 | -0.8143 | No |
| 12 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 31736 | -0.972 | -0.8150 | No |
| 13 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 31758 | -0.979 | -0.8096 | No |
| 14 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 32153 | -1.113 | -0.8135 | No |
| 15 | CDC25C | cell division cycle 25C [Source:HGNC Symbol;Acc:HGNC:1727] | 32283 | -1.166 | -0.8098 | No |
| 16 | CENPA | centromere protein A [Source:HGNC Symbol;Acc:HGNC:1851] | 32379 | -1.207 | -0.8051 | No |
| 17 | POLD3 | DNA polymerase delta 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:20932] | 32927 | -1.435 | -0.8111 | No |
| 18 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 33403 | -1.555 | -0.8145 | No |
| 19 | CKS1B | CDC28 protein kinase regulatory subunit 1B [Source:HGNC Symbol;Acc:HGNC:19083] | 33975 | -1.818 | -0.8189 | Yes |
| 20 | PLOD2 | procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 [Source:HGNC Symbol;Acc:HGNC:9082] | 33994 | -1.825 | -0.8082 | Yes |
| 21 | BRD8 | bromodomain containing 8 [Source:HGNC Symbol;Acc:HGNC:19874] | 34042 | -1.851 | -0.7983 | Yes |
| 22 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34046 | -1.854 | -0.7871 | Yes |
| 23 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 34181 | -1.920 | -0.7790 | Yes |
| 24 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34248 | -1.957 | -0.7689 | Yes |
| 25 | CHAF1A | chromatin assembly factor 1 subunit A [Source:HGNC Symbol;Acc:HGNC:1910] | 34259 | -1.967 | -0.7572 | Yes |
| 26 | CCNB1 | cyclin B1 [Source:HGNC Symbol;Acc:HGNC:1579] | 34381 | -2.017 | -0.7482 | Yes |
| 27 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 34617 | -2.156 | -0.7414 | Yes |
| 28 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 34638 | -2.171 | -0.7288 | Yes |
| 29 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 34800 | -2.282 | -0.7192 | Yes |
| 30 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 34811 | -2.286 | -0.7056 | Yes |
| 31 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 34817 | -2.289 | -0.6918 | Yes |
| 32 | RAD51C | RAD51 paralog C [Source:HGNC Symbol;Acc:HGNC:9820] | 34859 | -2.317 | -0.6788 | Yes |
| 33 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 34921 | -2.364 | -0.6661 | Yes |
| 34 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 34947 | -2.382 | -0.6523 | Yes |
| 35 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35054 | -2.393 | -0.6406 | Yes |
| 36 | STIL | STIL centriolar assembly protein [Source:HGNC Symbol;Acc:HGNC:10879] | 35121 | -2.440 | -0.6275 | Yes |
| 37 | CDKN3 | cyclin dependent kinase inhibitor 3 [Source:HGNC Symbol;Acc:HGNC:1791] | 35122 | -2.440 | -0.6127 | Yes |
| 38 | CHEK1 | checkpoint kinase 1 [Source:HGNC Symbol;Acc:HGNC:1925] | 35152 | -2.473 | -0.5984 | Yes |
| 39 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 35217 | -2.531 | -0.5848 | Yes |
| 40 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 35484 | -2.727 | -0.5754 | Yes |
| 41 | CDC6 | cell division cycle 6 [Source:HGNC Symbol;Acc:HGNC:1744] | 35504 | -2.750 | -0.5591 | Yes |
| 42 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 35524 | -2.765 | -0.5428 | Yes |
| 43 | SPAG5 | sperm associated antigen 5 [Source:HGNC Symbol;Acc:HGNC:13452] | 35557 | -2.803 | -0.5267 | Yes |
| 44 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 35559 | -2.806 | -0.5096 | Yes |
| 45 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 35595 | -2.845 | -0.4933 | Yes |
| 46 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 35624 | -2.879 | -0.4765 | Yes |
| 47 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35635 | -2.889 | -0.4592 | Yes |
| 48 | GPX4 | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 35711 | -2.911 | -0.4435 | Yes |
| 49 | GALE | UDP-galactose-4-epimerase [Source:HGNC Symbol;Acc:HGNC:4116] | 35756 | -2.954 | -0.4267 | Yes |
| 50 | TRAIP | TRAF interacting protein [Source:HGNC Symbol;Acc:HGNC:30764] | 35797 | -2.987 | -0.4096 | Yes |
| 51 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 35809 | -2.998 | -0.3917 | Yes |
| 52 | OIP5 | Opa interacting protein 5 [Source:HGNC Symbol;Acc:HGNC:20300] | 35911 | -3.088 | -0.3757 | Yes |
| 53 | PLK1 | polo like kinase 1 [Source:HGNC Symbol;Acc:HGNC:9077] | 35969 | -3.152 | -0.3580 | Yes |
| 54 | SPC25 | SPC25 component of NDC80 kinetochore complex [Source:HGNC Symbol;Acc:HGNC:24031] | 36011 | -3.197 | -0.3397 | Yes |
| 55 | PTP4A3 | protein tyrosine phosphatase 4A3 [Source:HGNC Symbol;Acc:HGNC:9636] | 36043 | -3.241 | -0.3208 | Yes |
| 56 | KIF22 | kinesin family member 22 [Source:HGNC Symbol;Acc:HGNC:6391] | 36079 | -3.255 | -0.3020 | Yes |
| 57 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36104 | -3.290 | -0.2826 | Yes |
| 58 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 36135 | -3.335 | -0.2631 | Yes |
| 59 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 36211 | -3.412 | -0.2444 | Yes |
| 60 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36500 | -3.700 | -0.2297 | Yes |
| 61 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36503 | -3.702 | -0.2072 | Yes |
| 62 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36567 | -3.812 | -0.1857 | Yes |
| 63 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36580 | -3.830 | -0.1628 | Yes |
| 64 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36607 | -3.868 | -0.1399 | Yes |
| 65 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 36610 | -3.875 | -0.1164 | Yes |
| 66 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 36711 | -3.993 | -0.0948 | Yes |
| 67 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36726 | -4.014 | -0.0708 | Yes |
| 68 | WEE1 | WEE1 G2 checkpoint kinase [Source:HGNC Symbol;Acc:HGNC:12761] | 36881 | -4.323 | -0.0486 | Yes |
| 69 | CHEK2 | checkpoint kinase 2 [Source:HGNC Symbol;Acc:HGNC:16627] | 36935 | -4.445 | -0.0230 | Yes |
| 70 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 36948 | -4.472 | 0.0038 | Yes |