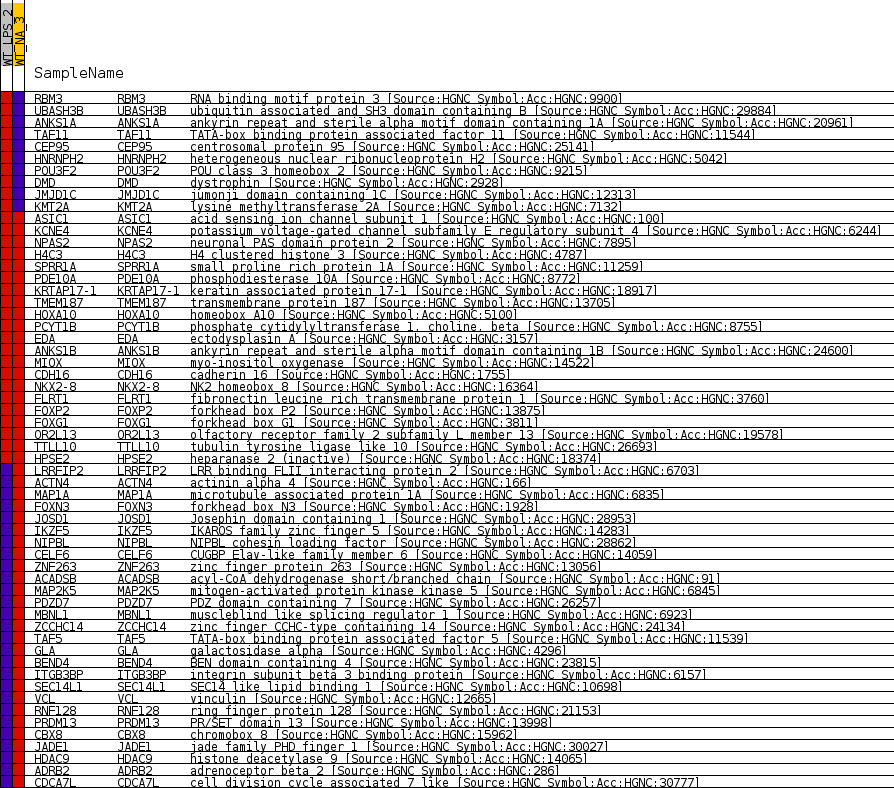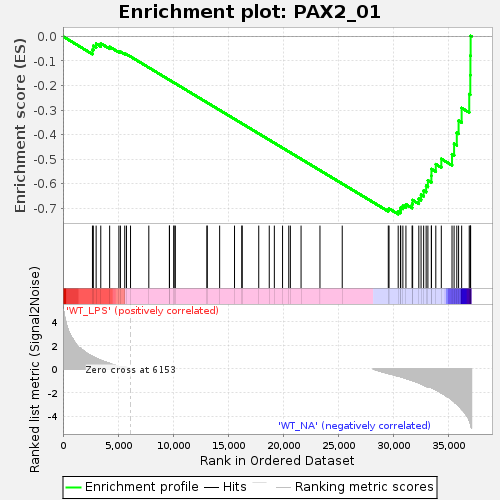
Fig 1: Enrichment plot: PAX2_01
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #WT_LPS_versus_WT_NA.Nof1.cls #WT_LPS_versus_WT_NA_repos |
| Phenotype | Nof1.cls#WT_LPS_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | PAX2_01 |
| Enrichment Score (ES) | -0.7243831 |
| Normalized Enrichment Score (NES) | -1.4286687 |
| Nominal p-value | 0.008739077 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RBM3 | RNA binding motif protein 3 [Source:HGNC Symbol;Acc:HGNC:9900] | 2674 | 1.047 | -0.0540 | No |
| 2 | UBASH3B | ubiquitin associated and SH3 domain containing B [Source:HGNC Symbol;Acc:HGNC:29884] | 2752 | 1.010 | -0.0384 | No |
| 3 | ANKS1A | ankyrin repeat and sterile alpha motif domain containing 1A [Source:HGNC Symbol;Acc:HGNC:20961] | 3008 | 0.892 | -0.0298 | No |
| 4 | TAF11 | TATA-box binding protein associated factor 11 [Source:HGNC Symbol;Acc:HGNC:11544] | 3429 | 0.728 | -0.0284 | No |
| 5 | CEP95 | centrosomal protein 95 [Source:HGNC Symbol;Acc:HGNC:25141] | 4235 | 0.452 | -0.0423 | No |
| 6 | HNRNPH2 | heterogeneous nuclear ribonucleoprotein H2 [Source:HGNC Symbol;Acc:HGNC:5042] | 5074 | 0.206 | -0.0613 | No |
| 7 | POU3F2 | POU class 3 homeobox 2 [Source:HGNC Symbol;Acc:HGNC:9215] | 5211 | 0.169 | -0.0621 | No |
| 8 | DMD | dystrophin [Source:HGNC Symbol;Acc:HGNC:2928] | 5605 | 0.153 | -0.0700 | No |
| 9 | JMJD1C | jumonji domain containing 1C [Source:HGNC Symbol;Acc:HGNC:12313] | 5761 | 0.115 | -0.0722 | No |
| 10 | KMT2A | lysine methyltransferase 2A [Source:HGNC Symbol;Acc:HGNC:7132] | 6130 | 0.004 | -0.0821 | No |
| 11 | ASIC1 | acid sensing ion channel subunit 1 [Source:HGNC Symbol;Acc:HGNC:100] | 7790 | 0.000 | -0.1269 | No |
| 12 | KCNE4 | potassium voltage-gated channel subfamily E regulatory subunit 4 [Source:HGNC Symbol;Acc:HGNC:6244] | 9660 | 0.000 | -0.1773 | No |
| 13 | NPAS2 | neuronal PAS domain protein 2 [Source:HGNC Symbol;Acc:HGNC:7895] | 9664 | 0.000 | -0.1774 | No |
| 14 | H4C3 | H4 clustered histone 3 [Source:HGNC Symbol;Acc:HGNC:4787] | 10055 | 0.000 | -0.1880 | No |
| 15 | SPRR1A | small proline rich protein 1A [Source:HGNC Symbol;Acc:HGNC:11259] | 10085 | 0.000 | -0.1887 | No |
| 16 | PDE10A | phosphodiesterase 10A [Source:HGNC Symbol;Acc:HGNC:8772] | 10198 | 0.000 | -0.1918 | No |
| 17 | KRTAP17-1 | keratin associated protein 17-1 [Source:HGNC Symbol;Acc:HGNC:18917] | 13066 | 0.000 | -0.2692 | No |
| 18 | TMEM187 | transmembrane protein 187 [Source:HGNC Symbol;Acc:HGNC:13705] | 13091 | 0.000 | -0.2698 | No |
| 19 | HOXA10 | homeobox A10 [Source:HGNC Symbol;Acc:HGNC:5100] | 14220 | 0.000 | -0.3003 | No |
| 20 | PCYT1B | phosphate cytidylyltransferase 1, choline, beta [Source:HGNC Symbol;Acc:HGNC:8755] | 15570 | 0.000 | -0.3367 | No |
| 21 | EDA | ectodysplasin A [Source:HGNC Symbol;Acc:HGNC:3157] | 16230 | 0.000 | -0.3545 | No |
| 22 | ANKS1B | ankyrin repeat and sterile alpha motif domain containing 1B [Source:HGNC Symbol;Acc:HGNC:24600] | 16267 | 0.000 | -0.3555 | No |
| 23 | MIOX | myo-inositol oxygenase [Source:HGNC Symbol;Acc:HGNC:14522] | 17769 | 0.000 | -0.3960 | No |
| 24 | CDH16 | cadherin 16 [Source:HGNC Symbol;Acc:HGNC:1755] | 18723 | 0.000 | -0.4217 | No |
| 25 | NKX2-8 | NK2 homeobox 8 [Source:HGNC Symbol;Acc:HGNC:16364] | 19186 | 0.000 | -0.4342 | No |
| 26 | FLRT1 | fibronectin leucine rich transmembrane protein 1 [Source:HGNC Symbol;Acc:HGNC:3760] | 19931 | 0.000 | -0.4543 | No |
| 27 | FOXP2 | forkhead box P2 [Source:HGNC Symbol;Acc:HGNC:13875] | 20495 | 0.000 | -0.4695 | No |
| 28 | FOXG1 | forkhead box G1 [Source:HGNC Symbol;Acc:HGNC:3811] | 20641 | 0.000 | -0.4734 | No |
| 29 | OR2L13 | olfactory receptor family 2 subfamily L member 13 [Source:HGNC Symbol;Acc:HGNC:19578] | 21611 | 0.000 | -0.4996 | No |
| 30 | TTLL10 | tubulin tyrosine ligase like 10 [Source:HGNC Symbol;Acc:HGNC:26693] | 23327 | 0.000 | -0.5459 | No |
| 31 | HPSE2 | heparanase 2 (inactive) [Source:HGNC Symbol;Acc:HGNC:18374] | 25349 | 0.000 | -0.6005 | No |
| 32 | LRRFIP2 | LRR binding FLII interacting protein 2 [Source:HGNC Symbol;Acc:HGNC:6703] | 29529 | -0.370 | -0.7069 | No |
| 33 | ACTN4 | actinin alpha 4 [Source:HGNC Symbol;Acc:HGNC:166] | 29596 | -0.387 | -0.7019 | No |
| 34 | MAP1A | microtubule associated protein 1A [Source:HGNC Symbol;Acc:HGNC:6835] | 30429 | -0.588 | -0.7141 | Yes |
| 35 | FOXN3 | forkhead box N3 [Source:HGNC Symbol;Acc:HGNC:1928] | 30634 | -0.642 | -0.7085 | Yes |
| 36 | JOSD1 | Josephin domain containing 1 [Source:HGNC Symbol;Acc:HGNC:28953] | 30658 | -0.648 | -0.6978 | Yes |
| 37 | IKZF5 | IKAROS family zinc finger 5 [Source:HGNC Symbol;Acc:HGNC:14283] | 30854 | -0.697 | -0.6909 | Yes |
| 38 | NIPBL | NIPBL cohesin loading factor [Source:HGNC Symbol;Acc:HGNC:28862] | 31131 | -0.793 | -0.6846 | Yes |
| 39 | CELF6 | CUGBP Elav-like family member 6 [Source:HGNC Symbol;Acc:HGNC:14059] | 31692 | -0.961 | -0.6829 | Yes |
| 40 | ZNF263 | zinc finger protein 263 [Source:HGNC Symbol;Acc:HGNC:13056] | 31740 | -0.974 | -0.6672 | Yes |
| 41 | ACADSB | acyl-CoA dehydrogenase short/branched chain [Source:HGNC Symbol;Acc:HGNC:91] | 32301 | -1.176 | -0.6619 | Yes |
| 42 | MAP2K5 | mitogen-activated protein kinase kinase 5 [Source:HGNC Symbol;Acc:HGNC:6845] | 32505 | -1.254 | -0.6455 | Yes |
| 43 | PDZD7 | PDZ domain containing 7 [Source:HGNC Symbol;Acc:HGNC:26257] | 32740 | -1.345 | -0.6284 | Yes |
| 44 | MBNL1 | muscleblind like splicing regulator 1 [Source:HGNC Symbol;Acc:HGNC:6923] | 32966 | -1.450 | -0.6092 | Yes |
| 45 | ZCCHC14 | zinc finger CCHC-type containing 14 [Source:HGNC Symbol;Acc:HGNC:24134] | 33125 | -1.518 | -0.5870 | Yes |
| 46 | TAF5 | TATA-box binding protein associated factor 5 [Source:HGNC Symbol;Acc:HGNC:11539] | 33441 | -1.573 | -0.5681 | Yes |
| 47 | GLA | galactosidase alpha [Source:HGNC Symbol;Acc:HGNC:4296] | 33446 | -1.575 | -0.5408 | Yes |
| 48 | BEND4 | BEN domain containing 4 [Source:HGNC Symbol;Acc:HGNC:23815] | 33843 | -1.753 | -0.5209 | Yes |
| 49 | ITGB3BP | integrin subunit beta 3 binding protein [Source:HGNC Symbol;Acc:HGNC:6157] | 34343 | -2.014 | -0.4993 | Yes |
| 50 | SEC14L1 | SEC14 like lipid binding 1 [Source:HGNC Symbol;Acc:HGNC:10698] | 35315 | -2.598 | -0.4803 | Yes |
| 51 | VCL | vinculin [Source:HGNC Symbol;Acc:HGNC:12665] | 35503 | -2.750 | -0.4374 | Yes |
| 52 | RNF128 | ring finger protein 128 [Source:HGNC Symbol;Acc:HGNC:21153] | 35749 | -2.947 | -0.3927 | Yes |
| 53 | PRDM13 | PR/SET domain 13 [Source:HGNC Symbol;Acc:HGNC:13998] | 35912 | -3.089 | -0.3432 | Yes |
| 54 | CBX8 | chromobox 8 [Source:HGNC Symbol;Acc:HGNC:15962] | 36189 | -3.383 | -0.2917 | Yes |
| 55 | JADE1 | jade family PHD finger 1 [Source:HGNC Symbol;Acc:HGNC:30027] | 36878 | -4.310 | -0.2352 | Yes |
| 56 | HDAC9 | histone deacetylase 9 [Source:HGNC Symbol;Acc:HGNC:14065] | 36974 | -4.567 | -0.1582 | Yes |
| 57 | ADRB2 | adrenoceptor beta 2 [Source:HGNC Symbol;Acc:HGNC:286] | 36985 | -4.601 | -0.0783 | Yes |
| 58 | CDCA7L | cell division cycle associated 7 like [Source:HGNC Symbol;Acc:HGNC:30777] | 37005 | -4.656 | 0.0023 | Yes |