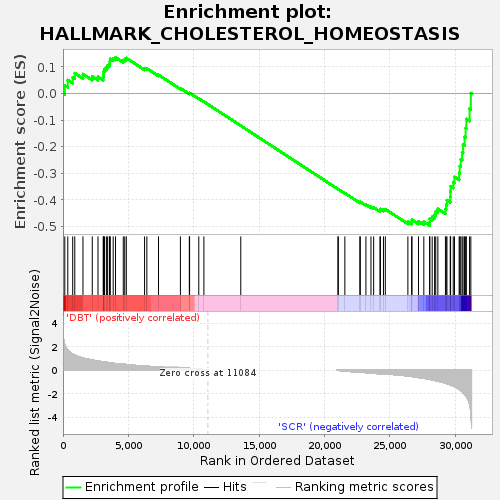
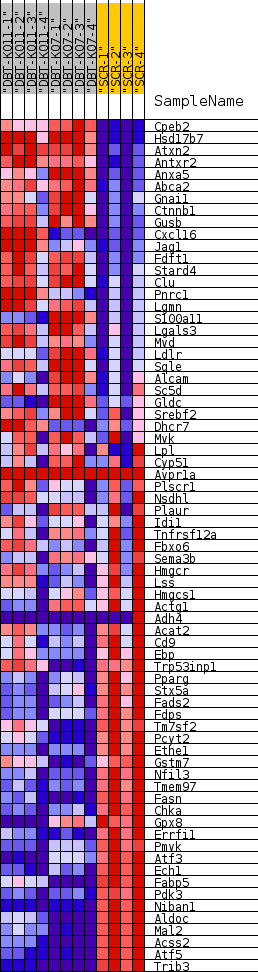
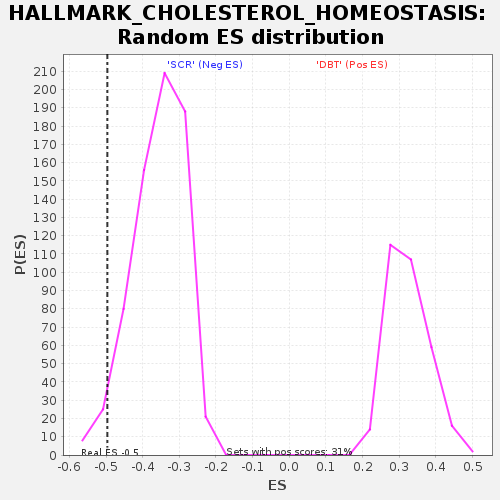
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | Cpeb2 | cytoplasmic polyadenylation element binding protein 2 [Source:MGI Symbol;Acc:MGI:2442640] | 137 | 2.144 | 0.0306 | No |
| 2 | Hsd17b7 | hydroxysteroid (17-beta) dehydrogenase 7 [Source:MGI Symbol;Acc:MGI:1330808] | 366 | 1.685 | 0.0508 | No |
| 3 | Atxn2 | ataxin 2 [Source:MGI Symbol;Acc:MGI:1277223] | 741 | 1.354 | 0.0609 | No |
| 4 | Antxr2 | anthrax toxin receptor 2 [Source:MGI Symbol;Acc:MGI:1919164] | 900 | 1.271 | 0.0766 | No |
| 5 | Anxa5 | annexin A5 [Source:MGI Symbol;Acc:MGI:106008] | 1526 | 1.025 | 0.0732 | No |
| 6 | Abca2 | ATP-binding cassette, sub-family A (ABC1), member 2 [Source:MGI Symbol;Acc:MGI:99606] | 2237 | 0.859 | 0.0645 | No |
| 7 | Gnai1 | guanine nucleotide binding protein (G protein), alpha inhibiting 1 [Source:MGI Symbol;Acc:MGI:95771] | 2676 | 0.769 | 0.0630 | No |
| 8 | Ctnnb1 | catenin (cadherin associated protein), beta 1 [Source:MGI Symbol;Acc:MGI:88276] | 3073 | 0.698 | 0.0617 | No |
| 9 | Gusb | glucuronidase, beta [Source:MGI Symbol;Acc:MGI:95872] | 3096 | 0.693 | 0.0723 | No |
| 10 | Cxcl16 | chemokine (C-X-C motif) ligand 16 [Source:MGI Symbol;Acc:MGI:1932682] | 3106 | 0.691 | 0.0833 | No |
| 11 | Jag1 | jagged 1 [Source:MGI Symbol;Acc:MGI:1095416] | 3176 | 0.683 | 0.0922 | No |
| 12 | Fdft1 | farnesyl diphosphate farnesyl transferase 1 [Source:MGI Symbol;Acc:MGI:102706] | 3317 | 0.662 | 0.0985 | No |
| 13 | Stard4 | StAR-related lipid transfer (START) domain containing 4 [Source:MGI Symbol;Acc:MGI:2156764] | 3410 | 0.654 | 0.1062 | No |
| 14 | Clu | clusterin [Source:MGI Symbol;Acc:MGI:88423] | 3550 | 0.626 | 0.1120 | No |
| 15 | Pnrc1 | proline-rich nuclear receptor coactivator 1 [Source:MGI Symbol;Acc:MGI:1917838] | 3578 | 0.622 | 0.1213 | No |
| 16 | Lgmn | legumain [Source:MGI Symbol;Acc:MGI:1330838] | 3608 | 0.616 | 0.1304 | No |
| 17 | S100a11 | S100 calcium binding protein A11 [Source:MGI Symbol;Acc:MGI:1338798] | 3838 | 0.580 | 0.1325 | No |
| 18 | Lgals3 | lectin, galactose binding, soluble 3 [Source:MGI Symbol;Acc:MGI:96778] | 4015 | 0.557 | 0.1359 | No |
| 19 | Mvd | mevalonate (diphospho) decarboxylase [Source:MGI Symbol;Acc:MGI:2179327] | 4608 | 0.504 | 0.1252 | No |
| 20 | Ldlr | low density lipoprotein receptor [Source:MGI Symbol;Acc:MGI:96765] | 4720 | 0.490 | 0.1296 | No |
| 21 | Sqle | squalene epoxidase [Source:MGI Symbol;Acc:MGI:109296] | 4840 | 0.475 | 0.1336 | No |
| 22 | Alcam | activated leukocyte cell adhesion molecule [Source:MGI Symbol;Acc:MGI:1313266] | 6236 | 0.359 | 0.0947 | No |
| 23 | Sc5d | sterol-C5-desaturase [Source:MGI Symbol;Acc:MGI:1353611] | 6411 | 0.337 | 0.0946 | No |
| 24 | Gldc | glycine decarboxylase [Source:MGI Symbol;Acc:MGI:1341155] | 7302 | 0.265 | 0.0703 | No |
| 25 | Srebf2 | sterol regulatory element binding factor 2 [Source:MGI Symbol;Acc:MGI:107585] | 8976 | 0.217 | 0.0202 | No |
| 26 | Dhcr7 | 7-dehydrocholesterol reductase [Source:MGI Symbol;Acc:MGI:1298378] | 9658 | 0.148 | 0.0008 | No |
| 27 | Mvk | mevalonate kinase [Source:MGI Symbol;Acc:MGI:107624] | 9679 | 0.146 | 0.0025 | No |
| 28 | Lpl | lipoprotein lipase [Source:MGI Symbol;Acc:MGI:96820] | 10381 | 0.083 | -0.0186 | No |
| 29 | Cyp51 | cytochrome P450, family 51 [Source:MGI Symbol;Acc:MGI:106040] | 10772 | 0.044 | -0.0304 | No |
| 30 | Avpr1a | arginine vasopressin receptor 1A [Source:MGI Symbol;Acc:MGI:1859216] | 13592 | 0.000 | -0.1209 | No |
| 31 | Plscr1 | phospholipid scramblase 1 [Source:MGI Symbol;Acc:MGI:893575] | 21020 | -0.012 | -0.3590 | No |
| 32 | Nsdhl | NAD(P) dependent steroid dehydrogenase-like [Source:MGI Symbol;Acc:MGI:1099438] | 21050 | -0.018 | -0.3596 | No |
| 33 | Plaur | plasminogen activator, urokinase receptor [Source:MGI Symbol;Acc:MGI:97612] | 21549 | -0.074 | -0.3744 | No |
| 34 | Idi1 | isopentenyl-diphosphate delta isomerase [Source:MGI Symbol;Acc:MGI:2442264] | 22696 | -0.158 | -0.4086 | No |
| 35 | Tnfrsf12a | tumor necrosis factor receptor superfamily, member 12a [Source:MGI Symbol;Acc:MGI:1351484] | 22729 | -0.163 | -0.4069 | No |
| 36 | Fbxo6 | F-box protein 6 [Source:MGI Symbol;Acc:MGI:1354743] | 23160 | -0.211 | -0.4173 | No |
| 37 | Sema3b | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B [Source:MGI Symbol;Acc:MGI:107561] | 23546 | -0.246 | -0.4256 | No |
| 38 | Hmgcr | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase [Source:MGI Symbol;Acc:MGI:96159] | 23752 | -0.262 | -0.4279 | No |
| 39 | Lss | lanosterol synthase [Source:MGI Symbol;Acc:MGI:1336155] | 24235 | -0.313 | -0.4383 | No |
| 40 | Hmgcs1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 [Source:MGI Symbol;Acc:MGI:107592] | 24267 | -0.317 | -0.4341 | No |
| 41 | Actg1 | actin, gamma, cytoplasmic 1 [Source:MGI Symbol;Acc:MGI:87906] | 24501 | -0.346 | -0.4359 | No |
| 42 | Adh4 | alcohol dehydrogenase 4 (class II), pi polypeptide [Source:MGI Symbol;Acc:MGI:1349472] | 24643 | -0.357 | -0.4346 | No |
| 43 | Acat2 | acetyl-Coenzyme A acetyltransferase 2 [Source:MGI Symbol;Acc:MGI:87871] | 26368 | -0.492 | -0.4819 | No |
| 44 | Cd9 | CD9 antigen [Source:MGI Symbol;Acc:MGI:88348] | 26656 | -0.532 | -0.4824 | No |
| 45 | Ebp | phenylalkylamine Ca2+ antagonist (emopamil) binding protein [Source:MGI Symbol;Acc:MGI:107822] | 26681 | -0.538 | -0.4744 | No |
| 46 | Trp53inp1 | transformation related protein 53 inducible nuclear protein 1 [Source:MGI Symbol;Acc:MGI:1926609] | 27179 | -0.623 | -0.4802 | No |
| 47 | Pparg | peroxisome proliferator activated receptor gamma [Source:MGI Symbol;Acc:MGI:97747] | 27587 | -0.691 | -0.4820 | No |
| 48 | Stx5a | syntaxin 5A [Source:MGI Symbol;Acc:MGI:1928483] | 28034 | -0.780 | -0.4836 | Yes |
| 49 | Fads2 | fatty acid desaturase 2 [Source:MGI Symbol;Acc:MGI:1930079] | 28042 | -0.781 | -0.4710 | Yes |
| 50 | Fdps | farnesyl diphosphate synthetase [Source:MGI Symbol;Acc:MGI:104888] | 28227 | -0.823 | -0.4635 | Yes |
| 51 | Tm7sf2 | transmembrane 7 superfamily member 2 [Source:MGI Symbol;Acc:MGI:1920416] | 28411 | -0.868 | -0.4552 | Yes |
| 52 | Pcyt2 | phosphate cytidylyltransferase 2, ethanolamine [Source:MGI Symbol;Acc:MGI:1915921] | 28499 | -0.888 | -0.4435 | Yes |
| 53 | Ethe1 | ethylmalonic encephalopathy 1 [Source:MGI Symbol;Acc:MGI:1913321] | 28661 | -0.932 | -0.4335 | Yes |
| 54 | Gstm7 | glutathione S-transferase, mu 7 [Source:MGI Symbol;Acc:MGI:1915562] | 29239 | -1.101 | -0.4340 | Yes |
| 55 | Nfil3 | nuclear factor, interleukin 3, regulated [Source:MGI Symbol;Acc:MGI:109495] | 29296 | -1.121 | -0.4175 | Yes |
| 56 | Tmem97 | transmembrane protein 97 [Source:MGI Symbol;Acc:MGI:1916321] | 29355 | -1.141 | -0.4008 | Yes |
| 57 | Fasn | fatty acid synthase [Source:MGI Symbol;Acc:MGI:95485] | 29614 | -1.246 | -0.3887 | Yes |
| 58 | Chka | choline kinase alpha [Source:MGI Symbol;Acc:MGI:107760] | 29619 | -1.248 | -0.3685 | Yes |
| 59 | Gpx8 | glutathione peroxidase 8 (putative) [Source:MGI Symbol;Acc:MGI:1916840] | 29635 | -1.255 | -0.3485 | Yes |
| 60 | Errfi1 | ERBB receptor feedback inhibitor 1 [Source:MGI Symbol;Acc:MGI:1921405] | 29836 | -1.341 | -0.3330 | Yes |
| 61 | Pmvk | phosphomevalonate kinase [Source:MGI Symbol;Acc:MGI:1915853] | 29931 | -1.391 | -0.3133 | Yes |
| 62 | Atf3 | activating transcription factor 3 [Source:MGI Symbol;Acc:MGI:109384] | 30287 | -1.613 | -0.2984 | Yes |
| 63 | Ech1 | enoyl coenzyme A hydratase 1, peroxisomal [Source:MGI Symbol;Acc:MGI:1858208] | 30334 | -1.652 | -0.2729 | Yes |
| 64 | Fabp5 | fatty acid binding protein 5, epidermal [Source:MGI Symbol;Acc:MGI:101790] | 30410 | -1.709 | -0.2474 | Yes |
| 65 | Pdk3 | pyruvate dehydrogenase kinase, isoenzyme 3 [Source:MGI Symbol;Acc:MGI:2384308] | 30516 | -1.813 | -0.2212 | Yes |
| 66 | Niban1 | niban apoptosis regulator 1 [Source:MGI Symbol;Acc:MGI:2137237] | 30586 | -1.899 | -0.1924 | Yes |
| 67 | Aldoc | aldolase C, fructose-bisphosphate [Source:MGI Symbol;Acc:MGI:101863] | 30700 | -2.040 | -0.1627 | Yes |
| 68 | Mal2 | mal, T cell differentiation protein 2 [Source:MGI Symbol;Acc:MGI:2146021] | 30782 | -2.145 | -0.1303 | Yes |
| 69 | Acss2 | acyl-CoA synthetase short-chain family member 2 [Source:MGI Symbol;Acc:MGI:1890410] | 30838 | -2.227 | -0.0957 | Yes |
| 70 | Atf5 | activating transcription factor 5 [Source:MGI Symbol;Acc:MGI:2141857] | 31085 | -2.899 | -0.0563 | Yes |
| 71 | Trib3 | tribbles pseudokinase 3 [Source:MGI Symbol;Acc:MGI:1345675] | 31182 | -3.750 | 0.0018 | Yes |
Table: GSEA details [plain text format]