| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | Pom121 | nuclear pore membrane protein 121 [Source:MGI Symbol;Acc:MGI:2137624] | 605 | 1.449 | -0.0027 | No |
| 2 | Stx3 | syntaxin 3 [Source:MGI Symbol;Acc:MGI:103077] | 974 | 1.234 | -0.0002 | No |
| 3 | Gtf2h1 | general transcription factor II H, polypeptide 1 [Source:MGI Symbol;Acc:MGI:1277216] | 983 | 1.228 | 0.0138 | No |
| 4 | Ncbp2 | nuclear cap binding protein subunit 2 [Source:MGI Symbol;Acc:MGI:1915342] | 1009 | 1.218 | 0.0271 | No |
| 5 | Gtf3c5 | general transcription factor IIIC, polypeptide 5 [Source:MGI Symbol;Acc:MGI:1917489] | 1013 | 1.216 | 0.0411 | No |
| 6 | Vps37b | vacuolar protein sorting 37B [Source:MGI Symbol;Acc:MGI:1916724] | 1316 | 1.099 | 0.0442 | No |
| 7 | Ada | adenosine deaminase [Source:MGI Symbol;Acc:MGI:87916] | 1476 | 1.043 | 0.0512 | No |
| 8 | Adcy6 | adenylate cyclase 6 [Source:MGI Symbol;Acc:MGI:87917] | 1518 | 1.026 | 0.0618 | No |
| 9 | Npr2 | natriuretic peptide receptor 2 [Source:MGI Symbol;Acc:MGI:97372] | 1542 | 1.021 | 0.0729 | No |
| 10 | Dgcr8 | DGCR8, microprocessor complex subunit [Source:MGI Symbol;Acc:MGI:2151114] | 1554 | 1.016 | 0.0843 | No |
| 11 | Sac3d1 | SAC3 domain containing 1 [Source:MGI Symbol;Acc:MGI:1913656] | 1592 | 1.007 | 0.0948 | No |
| 12 | Snapc4 | small nuclear RNA activating complex, polypeptide 4 [Source:MGI Symbol;Acc:MGI:2443935] | 1713 | 0.976 | 0.1022 | No |
| 13 | Eloa | elongin A [Source:MGI Symbol;Acc:MGI:1351315] | 1965 | 0.914 | 0.1048 | No |
| 14 | Ak1 | adenylate kinase 1 [Source:MGI Symbol;Acc:MGI:87977] | 2179 | 0.871 | 0.1080 | No |
| 15 | Itpa | inosine triphosphatase (nucleoside triphosphate pyrophosphatase) [Source:MGI Symbol;Acc:MGI:96622] | 2206 | 0.866 | 0.1172 | No |
| 16 | Nudt9 | nudix hydrolase 9 [Source:MGI Symbol;Acc:MGI:1921417] | 2425 | 0.821 | 0.1197 | No |
| 17 | Taf1c | TATA-box binding protein associated factor, RNA polymerase I, C [Source:MGI Symbol;Acc:MGI:109576] | 2605 | 0.784 | 0.1231 | No |
| 18 | Dad1 | defender against cell death 1 [Source:MGI Symbol;Acc:MGI:101912] | 2889 | 0.733 | 0.1225 | No |
| 19 | Ercc1 | excision repair cross-complementing rodent repair deficiency, complementation group 1 [Source:MGI Symbol;Acc:MGI:95412] | 2981 | 0.717 | 0.1279 | No |
| 20 | Dctn4 | dynactin 4 [Source:MGI Symbol;Acc:MGI:1914915] | 3164 | 0.685 | 0.1300 | No |
| 21 | Polh | polymerase (DNA directed), eta (RAD 30 related) [Source:MGI Symbol;Acc:MGI:1891457] | 3566 | 0.624 | 0.1243 | No |
| 22 | Tarbp2 | TARBP2, RISC loading complex RNA binding subunit [Source:MGI Symbol;Acc:MGI:103027] | 3751 | 0.593 | 0.1253 | No |
| 23 | Ssrp1 | structure specific recognition protein 1 [Source:MGI Symbol;Acc:MGI:107912] | 3857 | 0.577 | 0.1286 | No |
| 24 | Upf3b | UPF3 regulator of nonsense transcripts homolog B (yeast) [Source:MGI Symbol;Acc:MGI:1915384] | 3924 | 0.567 | 0.1330 | No |
| 25 | Pola2 | polymerase (DNA directed), alpha 2 [Source:MGI Symbol;Acc:MGI:99690] | 3954 | 0.563 | 0.1386 | No |
| 26 | Pcna | proliferating cell nuclear antigen [Source:MGI Symbol;Acc:MGI:97503] | 4236 | 0.527 | 0.1357 | No |
| 27 | Ercc5 | excision repair cross-complementing rodent repair deficiency, complementation group 5 [Source:MGI Symbol;Acc:MGI:103582] | 4477 | 0.519 | 0.1340 | No |
| 28 | Ercc3 | excision repair cross-complementing rodent repair deficiency, complementation group 3 [Source:MGI Symbol;Acc:MGI:95414] | 4536 | 0.514 | 0.1381 | No |
| 29 | Nelfb | negative elongation factor complex member B [Source:MGI Symbol;Acc:MGI:1931035] | 4694 | 0.495 | 0.1388 | No |
| 30 | Fen1 | flap structure specific endonuclease 1 [Source:MGI Symbol;Acc:MGI:102779] | 4835 | 0.476 | 0.1398 | No |
| 31 | Ell | elongation factor RNA polymerase II [Source:MGI Symbol;Acc:MGI:109377] | 5261 | 0.420 | 0.1310 | No |
| 32 | Rae1 | ribonucleic acid export 1 [Source:MGI Symbol;Acc:MGI:1913929] | 5380 | 0.407 | 0.1320 | No |
| 33 | Pnp | purine-nucleoside phosphorylase [Source:MGI Symbol;Acc:MGI:97365] | 5628 | 0.393 | 0.1286 | No |
| 34 | Adrm1 | adhesion regulating molecule 1 26S proteasome ubiquitin receptor [Source:MGI Symbol;Acc:MGI:1929289] | 5677 | 0.388 | 0.1315 | No |
| 35 | Rpa2 | replication protein A2 [Source:MGI Symbol;Acc:MGI:1339939] | 5686 | 0.386 | 0.1357 | No |
| 36 | Ercc4 | excision repair cross-complementing rodent repair deficiency, complementation group 4 [Source:MGI Symbol;Acc:MGI:1354163] | 5761 | 0.378 | 0.1377 | No |
| 37 | Edf1 | endothelial differentiation-related factor 1 [Source:MGI Symbol;Acc:MGI:1891227] | 6121 | 0.369 | 0.1305 | No |
| 38 | Srsf6 | serine and arginine-rich splicing factor 6 [Source:MGI Symbol;Acc:MGI:1915246] | 6122 | 0.369 | 0.1348 | No |
| 39 | Tyms | thymidylate synthase [Source:MGI Symbol;Acc:MGI:98878] | 6123 | 0.369 | 0.1390 | No |
| 40 | Gtf2f1 | general transcription factor IIF, polypeptide 1 [Source:MGI Symbol;Acc:MGI:1923848] | 6194 | 0.363 | 0.1410 | No |
| 41 | Nelfcd | negative elongation factor complex member C/D, Th1l [Source:MGI Symbol;Acc:MGI:1926424] | 6347 | 0.345 | 0.1401 | No |
| 42 | Nfx1 | nuclear transcription factor, X-box binding 1 [Source:MGI Symbol;Acc:MGI:1921414] | 6432 | 0.335 | 0.1413 | No |
| 43 | Dut | deoxyuridine triphosphatase [Source:MGI Symbol;Acc:MGI:1346051] | 6590 | 0.317 | 0.1399 | No |
| 44 | Eif1b | eukaryotic translation initiation factor 1B [Source:MGI Symbol;Acc:MGI:1916219] | 6687 | 0.306 | 0.1404 | No |
| 45 | Ak3 | adenylate kinase 3 [Source:MGI Symbol;Acc:MGI:1860835] | 6918 | 0.289 | 0.1364 | No |
| 46 | Aprt | adenine phosphoribosyl transferase [Source:MGI Symbol;Acc:MGI:88061] | 6923 | 0.288 | 0.1396 | No |
| 47 | Surf1 | surfeit gene 1 [Source:MGI Symbol;Acc:MGI:98443] | 6953 | 0.284 | 0.1419 | No |
| 48 | Smad5 | SMAD family member 5 [Source:MGI Symbol;Acc:MGI:1328787] | 7439 | 0.249 | 0.1292 | No |
| 49 | Supt5 | suppressor of Ty 5, DSIF elongation factor subunit [Source:MGI Symbol;Acc:MGI:1202400] | 7497 | 0.242 | 0.1302 | No |
| 50 | Rad51 | RAD51 recombinase [Source:MGI Symbol;Acc:MGI:97890] | 7571 | 0.240 | 0.1306 | No |
| 51 | Clp1 | CLP1, cleavage and polyadenylation factor I subunit [Source:MGI Symbol;Acc:MGI:2138968] | 7584 | 0.239 | 0.1330 | No |
| 52 | Hcls1 | hematopoietic cell specific Lyn substrate 1 [Source:MGI Symbol;Acc:MGI:104568] | 8041 | 0.226 | 0.1210 | No |
| 53 | Gsdme | gasdermin E [Source:MGI Symbol;Acc:MGI:1889850] | 9218 | 0.197 | 0.0854 | No |
| 54 | Polr2a | polymerase (RNA) II (DNA directed) polypeptide A [Source:MGI Symbol;Acc:MGI:98086] | 9405 | 0.177 | 0.0815 | No |
| 55 | Cstf3 | cleavage stimulation factor, 3' pre-RNA, subunit 3 [Source:MGI Symbol;Acc:MGI:1351825] | 9581 | 0.157 | 0.0777 | No |
| 56 | Ccno | cyclin O [Source:MGI Symbol;Acc:MGI:2145534] | 9676 | 0.146 | 0.0764 | No |
| 57 | Polr2d | polymerase (RNA) II (DNA directed) polypeptide D [Source:MGI Symbol;Acc:MGI:1916491] | 10288 | 0.094 | 0.0578 | No |
| 58 | Ago4 | argonaute RISC catalytic subunit 4 [Source:MGI Symbol;Acc:MGI:1924100] | 10343 | 0.086 | 0.0571 | No |
| 59 | Rfc2 | replication factor C (activator 1) 2 [Source:MGI Symbol;Acc:MGI:1341868] | 10385 | 0.083 | 0.0567 | No |
| 60 | Zwint | ZW10 interactor [Source:MGI Symbol;Acc:MGI:1289227] | 10438 | 0.079 | 0.0560 | No |
| 61 | Sdcbp | syndecan binding protein [Source:MGI Symbol;Acc:MGI:1337026] | 10590 | 0.064 | 0.0518 | No |
| 62 | Gtf2h3 | general transcription factor IIH, polypeptide 3 [Source:MGI Symbol;Acc:MGI:1277143] | 10679 | 0.053 | 0.0496 | No |
| 63 | Rev3l | REV3 like, DNA directed polymerase zeta catalytic subunit [Source:MGI Symbol;Acc:MGI:1337131] | 10734 | 0.049 | 0.0485 | No |
| 64 | Sf3a3 | splicing factor 3a, subunit 3 [Source:MGI Symbol;Acc:MGI:1922312] | 10961 | 0.020 | 0.0414 | No |
| 65 | Ercc2 | excision repair cross-complementing rodent repair deficiency, complementation group 2 [Source:MGI Symbol;Acc:MGI:95413] | 10988 | 0.017 | 0.0408 | No |
| 66 | Usp11 | ubiquitin specific peptidase 11 [Source:MGI Symbol;Acc:MGI:2384312] | 21041 | -0.015 | -0.2824 | No |
| 67 | Trp53 | transformation related protein 53 [Source:MGI Symbol;Acc:MGI:98834] | 21201 | -0.037 | -0.2870 | No |
| 68 | Polr2i | polymerase (RNA) II (DNA directed) polypeptide I [Source:MGI Symbol;Acc:MGI:1917170] | 21332 | -0.050 | -0.2906 | No |
| 69 | Taf6 | TATA-box binding protein associated factor 6 [Source:MGI Symbol;Acc:MGI:109129] | 21372 | -0.054 | -0.2913 | No |
| 70 | Taf9 | TATA-box binding protein associated factor 9 [Source:MGI Symbol;Acc:MGI:1888697] | 21455 | -0.065 | -0.2932 | No |
| 71 | Umps | uridine monophosphate synthetase [Source:MGI Symbol;Acc:MGI:1298388] | 21626 | -0.081 | -0.2977 | No |
| 72 | Polr2j | polymerase (RNA) II (DNA directed) polypeptide J [Source:MGI Symbol;Acc:MGI:109582] | 21648 | -0.083 | -0.2974 | No |
| 73 | Vps37d | vacuolar protein sorting 37D [Source:MGI Symbol;Acc:MGI:2159402] | 21677 | -0.086 | -0.2973 | No |
| 74 | Rfc5 | replication factor C (activator 1) 5 [Source:MGI Symbol;Acc:MGI:1919401] | 21710 | -0.089 | -0.2973 | No |
| 75 | Alyref | Aly/REF export factor [Source:MGI Symbol;Acc:MGI:1341044] | 21804 | -0.097 | -0.2992 | No |
| 76 | Rala | v-ral simian leukemia viral oncogene A (ras related) [Source:MGI Symbol;Acc:MGI:1927243] | 21918 | -0.110 | -0.3015 | No |
| 77 | Brf2 | BRF2, RNA polymerase III transcription initiation factor 50kDa subunit [Source:MGI Symbol;Acc:MGI:1913903] | 22108 | -0.129 | -0.3061 | No |
| 78 | Pole4 | polymerase (DNA-directed), epsilon 4 (p12 subunit) [Source:MGI Symbol;Acc:MGI:1914229] | 22143 | -0.133 | -0.3056 | No |
| 79 | Nudt21 | nudix hydrolase 21 [Source:MGI Symbol;Acc:MGI:1915469] | 22226 | -0.143 | -0.3066 | No |
| 80 | Zfp707 | zinc finger protein 707 [Source:MGI Symbol;Acc:MGI:1916270] | 22644 | -0.150 | -0.3183 | No |
| 81 | Pde4b | phosphodiesterase 4B, cAMP specific [Source:MGI Symbol;Acc:MGI:99557] | 22652 | -0.151 | -0.3168 | No |
| 82 | Taf13 | TATA-box binding protein associated factor 13 [Source:MGI Symbol;Acc:MGI:1913500] | 22733 | -0.163 | -0.3175 | No |
| 83 | Gtf2b | general transcription factor IIB [Source:MGI Symbol;Acc:MGI:2385191] | 22853 | -0.172 | -0.3193 | No |
| 84 | Pde6g | phosphodiesterase 6G, cGMP-specific, rod, gamma [Source:MGI Symbol;Acc:MGI:97526] | 22980 | -0.188 | -0.3212 | No |
| 85 | Pola1 | polymerase (DNA directed), alpha 1 [Source:MGI Symbol;Acc:MGI:99660] | 23096 | -0.202 | -0.3225 | No |
| 86 | Polr1h | RNA polymerase I subunit H [Source:MGI Symbol;Acc:MGI:1913386] | 23099 | -0.202 | -0.3202 | No |
| 87 | Poll | polymerase (DNA directed), lambda [Source:MGI Symbol;Acc:MGI:1889000] | 23128 | -0.207 | -0.3187 | No |
| 88 | Cetn2 | centrin 2 [Source:MGI Symbol;Acc:MGI:1347085] | 23217 | -0.218 | -0.3190 | No |
| 89 | Polr2f | polymerase (RNA) II (DNA directed) polypeptide F [Source:MGI Symbol;Acc:MGI:1349393] | 23322 | -0.228 | -0.3197 | No |
| 90 | Sec61a1 | Sec61 alpha 1 subunit (S. cerevisiae) [Source:MGI Symbol;Acc:MGI:1858417] | 23406 | -0.237 | -0.3196 | No |
| 91 | Ddb1 | damage specific DNA binding protein 1 [Source:MGI Symbol;Acc:MGI:1202384] | 23559 | -0.247 | -0.3217 | No |
| 92 | Impdh2 | inosine monophosphate dehydrogenase 2 [Source:MGI Symbol;Acc:MGI:109367] | 23757 | -0.263 | -0.3249 | No |
| 93 | Pold1 | polymerase (DNA directed), delta 1, catalytic subunit [Source:MGI Symbol;Acc:MGI:97741] | 23855 | -0.275 | -0.3249 | No |
| 94 | Arl6ip1 | ADP-ribosylation factor-like 6 interacting protein 1 [Source:MGI Symbol;Acc:MGI:1858943] | 24045 | -0.291 | -0.3276 | No |
| 95 | Polr2h | polymerase (RNA) II (DNA directed) polypeptide H [Source:MGI Symbol;Acc:MGI:2384309] | 24207 | -0.310 | -0.3292 | No |
| 96 | Gmpr2 | guanosine monophosphate reductase 2 [Source:MGI Symbol;Acc:MGI:1917903] | 24360 | -0.328 | -0.3302 | No |
| 97 | Rfc3 | replication factor C (activator 1) 3 [Source:MGI Symbol;Acc:MGI:1916513] | 24464 | -0.340 | -0.3296 | No |
| 98 | Polr3c | polymerase (RNA) III (DNA directed) polypeptide C [Source:MGI Symbol;Acc:MGI:1921664] | 24568 | -0.353 | -0.3288 | No |
| 99 | Cmpk2 | cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial [Source:MGI Symbol;Acc:MGI:99830] | 24820 | -0.357 | -0.3328 | No |
| 100 | Cda | cytidine deaminase [Source:MGI Symbol;Acc:MGI:1919519] | 25291 | -0.375 | -0.3435 | Yes |
| 101 | Polr2c | polymerase (RNA) II (DNA directed) polypeptide C [Source:MGI Symbol;Acc:MGI:109299] | 25382 | -0.389 | -0.3419 | Yes |
| 102 | Nme1 | NME/NM23 nucleoside diphosphate kinase 1 [Source:MGI Symbol;Acc:MGI:97355] | 25405 | -0.392 | -0.3381 | Yes |
| 103 | Rbx1 | ring-box 1 [Source:MGI Symbol;Acc:MGI:1891829] | 25429 | -0.395 | -0.3342 | Yes |
| 104 | Lig1 | ligase I, DNA, ATP-dependent [Source:MGI Symbol;Acc:MGI:101789] | 25483 | -0.401 | -0.3313 | Yes |
| 105 | Bcap31 | B cell receptor associated protein 31 [Source:MGI Symbol;Acc:MGI:1350933] | 25645 | -0.411 | -0.3317 | Yes |
| 106 | Mrpl40 | mitochondrial ribosomal protein L40 [Source:MGI Symbol;Acc:MGI:1332635] | 25770 | -0.418 | -0.3308 | Yes |
| 107 | Rad52 | RAD52 homolog, DNA repair protein [Source:MGI Symbol;Acc:MGI:101949] | 26279 | -0.477 | -0.3416 | Yes |
| 108 | Polr2e | polymerase (RNA) II (DNA directed) polypeptide E [Source:MGI Symbol;Acc:MGI:1913670] | 26314 | -0.482 | -0.3371 | Yes |
| 109 | Tsg101 | tumor susceptibility gene 101 [Source:MGI Symbol;Acc:MGI:106581] | 26551 | -0.518 | -0.3387 | Yes |
| 110 | Polr1d | polymerase (RNA) I polypeptide D [Source:MGI Symbol;Acc:MGI:108403] | 26618 | -0.527 | -0.3347 | Yes |
| 111 | Ercc8 | excision repaiross-complementing rodent repair deficiency, complementation group 8 [Source:MGI Symbol;Acc:MGI:1919241] | 26649 | -0.531 | -0.3295 | Yes |
| 112 | Xpc | xeroderma pigmentosum, complementation group C [Source:MGI Symbol;Acc:MGI:103557] | 26811 | -0.558 | -0.3283 | Yes |
| 113 | Pold4 | polymerase (DNA-directed), delta 4 [Source:MGI Symbol;Acc:MGI:1916995] | 26868 | -0.569 | -0.3235 | Yes |
| 114 | Pold3 | polymerase (DNA-directed), delta 3, accessory subunit [Source:MGI Symbol;Acc:MGI:1915217] | 26883 | -0.572 | -0.3173 | Yes |
| 115 | Rnmt | RNA (guanine-7-) methyltransferase [Source:MGI Symbol;Acc:MGI:1915147] | 27171 | -0.621 | -0.3193 | Yes |
| 116 | Taf10 | TATA-box binding protein associated factor 10 [Source:MGI Symbol;Acc:MGI:1346320] | 27487 | -0.667 | -0.3217 | Yes |
| 117 | Cox17 | cytochrome c oxidase assembly protein 17, copper chaperone [Source:MGI Symbol;Acc:MGI:1333806] | 27663 | -0.705 | -0.3191 | Yes |
| 118 | Mpc2 | mitochondrial pyruvate carrier 2 [Source:MGI Symbol;Acc:MGI:1917706] | 27757 | -0.725 | -0.3137 | Yes |
| 119 | Bcam | basal cell adhesion molecule [Source:MGI Symbol;Acc:MGI:1929940] | 28043 | -0.781 | -0.3138 | Yes |
| 120 | Aaas | achalasia, adrenocortical insufficiency, alacrimia [Source:MGI Symbol;Acc:MGI:2443767] | 28070 | -0.788 | -0.3055 | Yes |
| 121 | Cant1 | calcium activated nucleotidase 1 [Source:MGI Symbol;Acc:MGI:1923275] | 28104 | -0.795 | -0.2974 | Yes |
| 122 | Gpx4 | glutathione peroxidase 4 [Source:MGI Symbol;Acc:MGI:104767] | 28361 | -0.856 | -0.2957 | Yes |
| 123 | Rfc4 | replication factor C (activator 1) 4 [Source:MGI Symbol;Acc:MGI:2146571] | 28415 | -0.868 | -0.2873 | Yes |
| 124 | Gtf2h5 | general transcription factor IIH, polypeptide 5 [Source:MGI Symbol;Acc:MGI:107227] | 28435 | -0.871 | -0.2778 | Yes |
| 125 | Nelfe | negative elongation factor complex member E, Rdbp [Source:MGI Symbol;Acc:MGI:102744] | 28451 | -0.876 | -0.2681 | Yes |
| 126 | Hprt | hypoxanthine guanine phosphoribosyl transferase [Source:MGI Symbol;Acc:MGI:96217] | 28469 | -0.879 | -0.2585 | Yes |
| 127 | Taf12 | TATA-box binding protein associated factor 12 [Source:MGI Symbol;Acc:MGI:1913714] | 28485 | -0.883 | -0.2487 | Yes |
| 128 | Polr2g | polymerase (RNA) II (DNA directed) polypeptide G [Source:MGI Symbol;Acc:MGI:1914960] | 28599 | -0.914 | -0.2417 | Yes |
| 129 | Polb | polymerase (DNA directed), beta [Source:MGI Symbol;Acc:MGI:97740] | 28644 | -0.929 | -0.2324 | Yes |
| 130 | Polr1c | polymerase (RNA) I polypeptide C [Source:MGI Symbol;Acc:MGI:103288] | 28785 | -0.965 | -0.2257 | Yes |
| 131 | Polr2k | polymerase (RNA) II (DNA directed) polypeptide K [Source:MGI Symbol;Acc:MGI:102725] | 29049 | -1.043 | -0.2220 | Yes |
| 132 | Prim1 | DNA primase, p49 subunit [Source:MGI Symbol;Acc:MGI:97757] | 29116 | -1.065 | -0.2118 | Yes |
| 133 | Nt5c3 | 5'-nucleotidase, cytosolic III [Source:MGI Symbol;Acc:MGI:1927186] | 29125 | -1.067 | -0.1997 | Yes |
| 134 | Gtf2a2 | general transcription factor II A, 2 [Source:MGI Symbol;Acc:MGI:1933289] | 29349 | -1.138 | -0.1936 | Yes |
| 135 | Tk2 | thymidine kinase 2, mitochondrial [Source:MGI Symbol;Acc:MGI:1913266] | 29358 | -1.142 | -0.1807 | Yes |
| 136 | Ddb2 | damage specific DNA binding protein 2 [Source:MGI Symbol;Acc:MGI:1355314] | 29374 | -1.149 | -0.1678 | Yes |
| 137 | Vps28 | vacuolar protein sorting 28 [Source:MGI Symbol;Acc:MGI:1914164] | 29489 | -1.195 | -0.1576 | Yes |
| 138 | Polr3gl | polymerase (RNA) III (DNA directed) polypeptide G like [Source:MGI Symbol;Acc:MGI:1917120] | 29719 | -1.290 | -0.1500 | Yes |
| 139 | Rpa3 | replication protein A3 [Source:MGI Symbol;Acc:MGI:1915490] | 29811 | -1.329 | -0.1375 | Yes |
| 140 | Snapc5 | small nuclear RNA activating complex, polypeptide 5 [Source:MGI Symbol;Acc:MGI:1914282] | 29864 | -1.354 | -0.1235 | Yes |
| 141 | Supt4a | SPT4A, DSIF elongation factor subunit [Source:MGI Symbol;Acc:MGI:107416] | 29925 | -1.386 | -0.1093 | Yes |
| 142 | Dguok | deoxyguanosine kinase [Source:MGI Symbol;Acc:MGI:1351602] | 29998 | -1.429 | -0.0951 | Yes |
| 143 | Nme3 | NME/NM23 nucleoside diphosphate kinase 3 [Source:MGI Symbol;Acc:MGI:1930182] | 30063 | -1.465 | -0.0801 | Yes |
| 144 | Rrm2b | ribonucleotide reductase M2 B (TP53 inducible) [Source:MGI Symbol;Acc:MGI:2155865] | 30526 | -1.832 | -0.0738 | Yes |
| 145 | Nt5c | 5',3'-nucleotidase, cytosolic [Source:MGI Symbol;Acc:MGI:1354954] | 30602 | -1.921 | -0.0539 | Yes |
| 146 | Mpg | N-methylpurine-DNA glycosylase [Source:MGI Symbol;Acc:MGI:97073] | 30738 | -2.082 | -0.0341 | Yes |
| 147 | Nme4 | NME/NM23 nucleoside diphosphate kinase 4 [Source:MGI Symbol;Acc:MGI:1931148] | 30790 | -2.157 | -0.0107 | Yes |
| 148 | Guk1 | guanylate kinase 1 [Source:MGI Symbol;Acc:MGI:95871] | 30794 | -2.162 | 0.0143 | Yes |
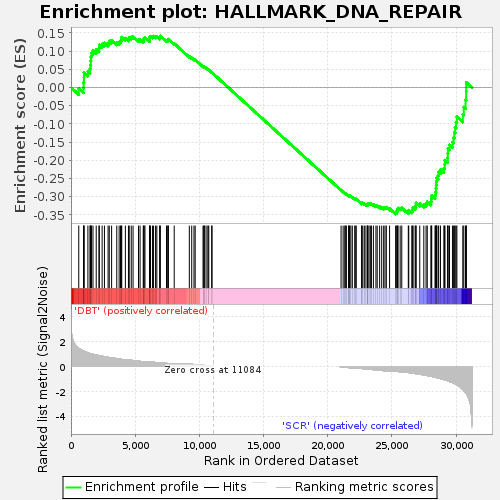
Table: GSEA details [plain text format]