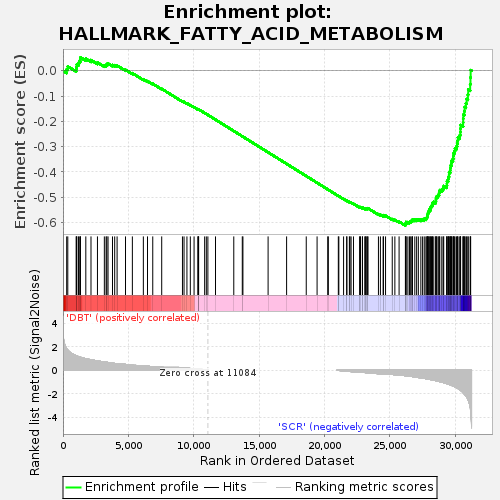
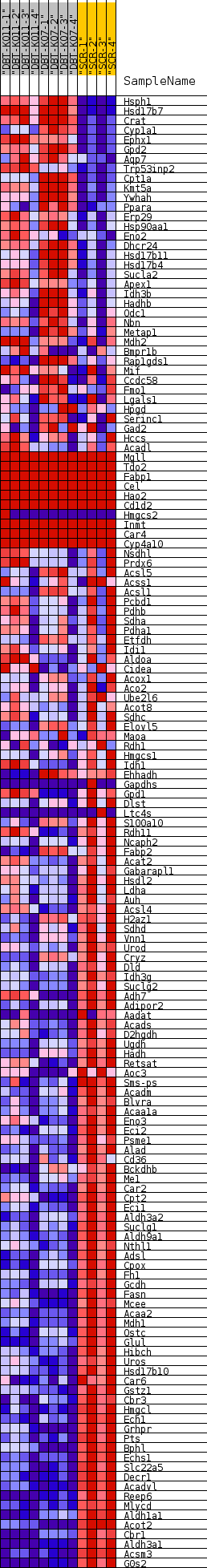
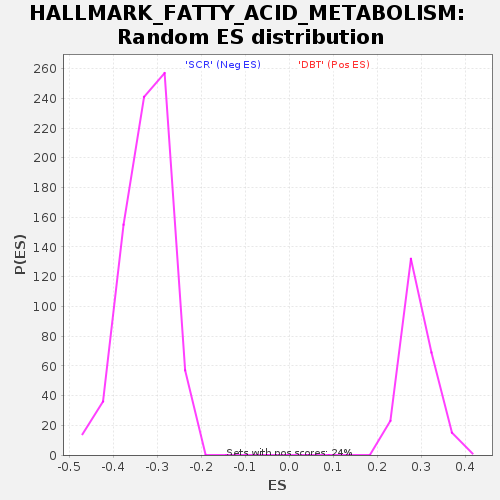
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | Hsph1 | heat shock 105kDa/110kDa protein 1 [Source:MGI Symbol;Acc:MGI:105053] | 282 | 1.809 | 0.0058 | No |
| 2 | Hsd17b7 | hydroxysteroid (17-beta) dehydrogenase 7 [Source:MGI Symbol;Acc:MGI:1330808] | 366 | 1.685 | 0.0171 | No |
| 3 | Crat | carnitine acetyltransferase [Source:MGI Symbol;Acc:MGI:109501] | 1005 | 1.219 | 0.0066 | No |
| 4 | Cyp1a1 | cytochrome P450, family 1, subfamily a, polypeptide 1 [Source:MGI Symbol;Acc:MGI:88588] | 1039 | 1.203 | 0.0154 | No |
| 5 | Ephx1 | epoxide hydrolase 1, microsomal [Source:MGI Symbol;Acc:MGI:95405] | 1054 | 1.194 | 0.0248 | No |
| 6 | Gpd2 | glycerol phosphate dehydrogenase 2, mitochondrial [Source:MGI Symbol;Acc:MGI:99778] | 1188 | 1.148 | 0.0300 | No |
| 7 | Aqp7 | aquaporin 7 [Source:MGI Symbol;Acc:MGI:1314647] | 1240 | 1.126 | 0.0377 | No |
| 8 | Trp53inp2 | transformation related protein 53 inducible nuclear protein 2 [Source:MGI Symbol;Acc:MGI:1915978] | 1329 | 1.094 | 0.0438 | No |
| 9 | Cpt1a | carnitine palmitoyltransferase 1a, liver [Source:MGI Symbol;Acc:MGI:1098296] | 1336 | 1.091 | 0.0526 | No |
| 10 | Kmt5a | lysine methyltransferase 5A [Source:MGI Symbol;Acc:MGI:1915206] | 1744 | 0.968 | 0.0475 | No |
| 11 | Ywhah | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide [Source:MGI Symbol;Acc:MGI:109194] | 2140 | 0.879 | 0.0421 | No |
| 12 | Ppara | peroxisome proliferator activated receptor alpha [Source:MGI Symbol;Acc:MGI:104740] | 2634 | 0.778 | 0.0326 | No |
| 13 | Erp29 | endoplasmic reticulum protein 29 [Source:MGI Symbol;Acc:MGI:1914647] | 3161 | 0.685 | 0.0213 | No |
| 14 | Hsp90aa1 | heat shock protein 90, alpha (cytosolic), class A member 1 [Source:MGI Symbol;Acc:MGI:96250] | 3295 | 0.666 | 0.0226 | No |
| 15 | Eno2 | enolase 2, gamma neuronal [Source:MGI Symbol;Acc:MGI:95394] | 3350 | 0.662 | 0.0263 | No |
| 16 | Dhcr24 | 24-dehydrocholesterol reductase [Source:MGI Symbol;Acc:MGI:1922004] | 3443 | 0.647 | 0.0287 | No |
| 17 | Hsd17b11 | hydroxysteroid (17-beta) dehydrogenase 11 [Source:MGI Symbol;Acc:MGI:2149821] | 3784 | 0.587 | 0.0226 | No |
| 18 | Hsd17b4 | hydroxysteroid (17-beta) dehydrogenase 4 [Source:MGI Symbol;Acc:MGI:105089] | 3952 | 0.563 | 0.0218 | No |
| 19 | Sucla2 | succinate-Coenzyme A ligase, ADP-forming, beta subunit [Source:MGI Symbol;Acc:MGI:1306775] | 4136 | 0.539 | 0.0204 | No |
| 20 | Apex1 | apurinic/apyrimidinic endonuclease 1 [Source:MGI Symbol;Acc:MGI:88042] | 4779 | 0.482 | 0.0037 | No |
| 21 | Idh3b | isocitrate dehydrogenase 3 (NAD+) beta [Source:MGI Symbol;Acc:MGI:2158650] | 5303 | 0.415 | -0.0097 | No |
| 22 | Hadhb | hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta [Source:MGI Symbol;Acc:MGI:2136381] | 6143 | 0.368 | -0.0337 | No |
| 23 | Odc1 | ornithine decarboxylase, structural 1 [Source:MGI Symbol;Acc:MGI:97402] | 6461 | 0.333 | -0.0411 | No |
| 24 | Nbn | nibrin [Source:MGI Symbol;Acc:MGI:1351625] | 6862 | 0.295 | -0.0515 | No |
| 25 | Metap1 | methionyl aminopeptidase 1 [Source:MGI Symbol;Acc:MGI:1922874] | 7547 | 0.241 | -0.0716 | No |
| 26 | Mdh2 | malate dehydrogenase 2, NAD (mitochondrial) [Source:MGI Symbol;Acc:MGI:97050] | 9133 | 0.199 | -0.1209 | No |
| 27 | Bmpr1b | bone morphogenetic protein receptor, type 1B [Source:MGI Symbol;Acc:MGI:107191] | 9254 | 0.193 | -0.1232 | No |
| 28 | Rap1gds1 | RAP1, GTP-GDP dissociation stimulator 1 [Source:MGI Symbol;Acc:MGI:2385189] | 9475 | 0.170 | -0.1289 | No |
| 29 | Mif | macrophage migration inhibitory factor (glycosylation-inhibiting factor) [Source:MGI Symbol;Acc:MGI:96982] | 9734 | 0.138 | -0.1360 | No |
| 30 | Ccdc58 | coiled-coil domain containing 58 [Source:MGI Symbol;Acc:MGI:2146423] | 10021 | 0.114 | -0.1443 | No |
| 31 | Fmo1 | flavin containing monooxygenase 1 [Source:MGI Symbol;Acc:MGI:1310002] | 10326 | 0.088 | -0.1533 | No |
| 32 | Lgals1 | lectin, galactose binding, soluble 1 [Source:MGI Symbol;Acc:MGI:96777] | 10336 | 0.087 | -0.1529 | No |
| 33 | Hpgd | hydroxyprostaglandin dehydrogenase 15 (NAD) [Source:MGI Symbol;Acc:MGI:108085] | 10339 | 0.087 | -0.1523 | No |
| 34 | Serinc1 | serine incorporator 1 [Source:MGI Symbol;Acc:MGI:1926228] | 10370 | 0.084 | -0.1525 | No |
| 35 | Gad2 | glutamic acid decarboxylase 2 [Source:MGI Symbol;Acc:MGI:95634] | 10830 | 0.037 | -0.1670 | No |
| 36 | Hccs | holocytochrome c synthetase [Source:MGI Symbol;Acc:MGI:106911] | 10957 | 0.021 | -0.1709 | No |
| 37 | Acadl | acyl-Coenzyme A dehydrogenase, long-chain [Source:MGI Symbol;Acc:MGI:87866] | 11077 | 0.002 | -0.1747 | No |
| 38 | Mgll | monoglyceride lipase [Source:MGI Symbol;Acc:MGI:1346042] | 11660 | 0.000 | -0.1934 | No |
| 39 | Tdo2 | tryptophan 2,3-dioxygenase [Source:MGI Symbol;Acc:MGI:1928486] | 13057 | 0.000 | -0.2383 | No |
| 40 | Fabp1 | fatty acid binding protein 1, liver [Source:MGI Symbol;Acc:MGI:95479] | 13707 | 0.000 | -0.2592 | No |
| 41 | Cel | carboxyl ester lipase [Source:MGI Symbol;Acc:MGI:88374] | 13756 | 0.000 | -0.2607 | No |
| 42 | Hao2 | hydroxyacid oxidase 2 [Source:MGI Symbol;Acc:MGI:96012] | 15681 | 0.000 | -0.3226 | No |
| 43 | Cd1d2 | CD1d2 antigen [Source:MGI Symbol;Acc:MGI:107675] | 17096 | 0.000 | -0.3681 | No |
| 44 | Hmgcs2 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 [Source:MGI Symbol;Acc:MGI:101939] | 18598 | 0.000 | -0.4164 | No |
| 45 | Inmt | indolethylamine N-methyltransferase [Source:MGI Symbol;Acc:MGI:102963] | 19426 | 0.000 | -0.4430 | No |
| 46 | Car4 | carbonic anhydrase 4 [Source:MGI Symbol;Acc:MGI:1096574] | 20241 | 0.000 | -0.4692 | No |
| 47 | Cyp4a10 | cytochrome P450, family 4, subfamily a, polypeptide 10 [Source:MGI Symbol;Acc:MGI:88611] | 20286 | 0.000 | -0.4706 | No |
| 48 | Nsdhl | NAD(P) dependent steroid dehydrogenase-like [Source:MGI Symbol;Acc:MGI:1099438] | 21050 | -0.018 | -0.4950 | No |
| 49 | Prdx6 | peroxiredoxin 6 [Source:MGI Symbol;Acc:MGI:894320] | 21089 | -0.023 | -0.4960 | No |
| 50 | Acsl5 | acyl-CoA synthetase long-chain family member 5 [Source:MGI Symbol;Acc:MGI:1919129] | 21452 | -0.064 | -0.5071 | No |
| 51 | Acss1 | acyl-CoA synthetase short-chain family member 1 [Source:MGI Symbol;Acc:MGI:1915988] | 21692 | -0.087 | -0.5141 | No |
| 52 | Acsl1 | acyl-CoA synthetase long-chain family member 1 [Source:MGI Symbol;Acc:MGI:102797] | 21702 | -0.088 | -0.5137 | No |
| 53 | Pcbd1 | pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 1 [Source:MGI Symbol;Acc:MGI:94873] | 21896 | -0.107 | -0.5190 | No |
| 54 | Pdhb | pyruvate dehydrogenase (lipoamide) beta [Source:MGI Symbol;Acc:MGI:1915513] | 21939 | -0.112 | -0.5194 | No |
| 55 | Sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) [Source:MGI Symbol;Acc:MGI:1914195] | 22026 | -0.119 | -0.5212 | No |
| 56 | Pdha1 | pyruvate dehydrogenase E1 alpha 1 [Source:MGI Symbol;Acc:MGI:97532] | 22209 | -0.141 | -0.5259 | No |
| 57 | Etfdh | electron transferring flavoprotein, dehydrogenase [Source:MGI Symbol;Acc:MGI:106100] | 22687 | -0.157 | -0.5400 | No |
| 58 | Idi1 | isopentenyl-diphosphate delta isomerase [Source:MGI Symbol;Acc:MGI:2442264] | 22696 | -0.158 | -0.5389 | No |
| 59 | Aldoa | aldolase A, fructose-bisphosphate [Source:MGI Symbol;Acc:MGI:87994] | 22747 | -0.164 | -0.5392 | No |
| 60 | Cidea | cell death-inducing DNA fragmentation factor, alpha subunit-like effector A [Source:MGI Symbol;Acc:MGI:1270845] | 22892 | -0.176 | -0.5424 | No |
| 61 | Acox1 | acyl-Coenzyme A oxidase 1, palmitoyl [Source:MGI Symbol;Acc:MGI:1330812] | 22897 | -0.177 | -0.5410 | No |
| 62 | Aco2 | aconitase 2, mitochondrial [Source:MGI Symbol;Acc:MGI:87880] | 23072 | -0.199 | -0.5450 | No |
| 63 | Ube2l6 | ubiquitin-conjugating enzyme E2L 6 [Source:MGI Symbol;Acc:MGI:1914500] | 23143 | -0.208 | -0.5455 | No |
| 64 | Acot8 | acyl-CoA thioesterase 8 [Source:MGI Symbol;Acc:MGI:2158201] | 23146 | -0.209 | -0.5439 | No |
| 65 | Sdhc | succinate dehydrogenase complex, subunit C, integral membrane protein [Source:MGI Symbol;Acc:MGI:1913302] | 23228 | -0.219 | -0.5447 | No |
| 66 | Elovl5 | ELOVL family member 5, elongation of long chain fatty acids (yeast) [Source:MGI Symbol;Acc:MGI:1916051] | 23296 | -0.226 | -0.5449 | No |
| 67 | Maoa | monoamine oxidase A [Source:MGI Symbol;Acc:MGI:96915] | 23311 | -0.227 | -0.5435 | No |
| 68 | Rdh1 | retinol dehydrogenase 1 (all trans) [Source:MGI Symbol;Acc:MGI:1195275] | 24117 | -0.300 | -0.5669 | No |
| 69 | Hmgcs1 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 [Source:MGI Symbol;Acc:MGI:107592] | 24267 | -0.317 | -0.5691 | No |
| 70 | Idh1 | isocitrate dehydrogenase 1 (NADP+), soluble [Source:MGI Symbol;Acc:MGI:96413] | 24474 | -0.341 | -0.5729 | No |
| 71 | Ehhadh | enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase [Source:MGI Symbol;Acc:MGI:1277964] | 24505 | -0.347 | -0.5710 | No |
| 72 | Gapdhs | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic [Source:MGI Symbol;Acc:MGI:95653] | 24661 | -0.357 | -0.5731 | No |
| 73 | Gpd1 | glycerol-3-phosphate dehydrogenase 1 (soluble) [Source:MGI Symbol;Acc:MGI:95679] | 25173 | -0.362 | -0.5865 | No |
| 74 | Dlst | dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) [Source:MGI Symbol;Acc:MGI:1926170] | 25373 | -0.388 | -0.5897 | No |
| 75 | Ltc4s | leukotriene C4 synthase [Source:MGI Symbol;Acc:MGI:107498] | 25696 | -0.417 | -0.5967 | No |
| 76 | S100a10 | S100 calcium binding protein A10 (calpactin) [Source:MGI Symbol;Acc:MGI:1339468] | 26178 | -0.467 | -0.6083 | Yes |
| 77 | Rdh11 | retinol dehydrogenase 11 [Source:MGI Symbol;Acc:MGI:102581] | 26187 | -0.468 | -0.6047 | Yes |
| 78 | Ncaph2 | non-SMC condensin II complex, subunit H2 [Source:MGI Symbol;Acc:MGI:1289164] | 26223 | -0.473 | -0.6019 | Yes |
| 79 | Fabp2 | fatty acid binding protein 2, intestinal [Source:MGI Symbol;Acc:MGI:95478] | 26228 | -0.474 | -0.5981 | Yes |
| 80 | Acat2 | acetyl-Coenzyme A acetyltransferase 2 [Source:MGI Symbol;Acc:MGI:87871] | 26368 | -0.492 | -0.5986 | Yes |
| 81 | Gabarapl1 | gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 [Source:MGI Symbol;Acc:MGI:1914980] | 26488 | -0.507 | -0.5982 | Yes |
| 82 | Hsdl2 | hydroxysteroid dehydrogenase like 2 [Source:MGI Symbol;Acc:MGI:1919729] | 26548 | -0.517 | -0.5958 | Yes |
| 83 | Ldha | lactate dehydrogenase A [Source:MGI Symbol;Acc:MGI:96759] | 26623 | -0.528 | -0.5939 | Yes |
| 84 | Auh | AU RNA binding protein/enoyl-coenzyme A hydratase [Source:MGI Symbol;Acc:MGI:1338011] | 26711 | -0.543 | -0.5922 | Yes |
| 85 | Acsl4 | acyl-CoA synthetase long-chain family member 4 [Source:MGI Symbol;Acc:MGI:1354713] | 26722 | -0.544 | -0.5880 | Yes |
| 86 | H2az1 | H2A.Z variant histone 1 [Source:MGI Symbol;Acc:MGI:1888388] | 26898 | -0.576 | -0.5889 | Yes |
| 87 | Sdhd | succinate dehydrogenase complex, subunit D, integral membrane protein [Source:MGI Symbol;Acc:MGI:1914175] | 27038 | -0.599 | -0.5885 | Yes |
| 88 | Vnn1 | vanin 1 [Source:MGI Symbol;Acc:MGI:108395] | 27168 | -0.620 | -0.5875 | Yes |
| 89 | Urod | uroporphyrinogen decarboxylase [Source:MGI Symbol;Acc:MGI:98916] | 27360 | -0.646 | -0.5883 | Yes |
| 90 | Cryz | crystallin, zeta [Source:MGI Symbol;Acc:MGI:88527] | 27494 | -0.669 | -0.5871 | Yes |
| 91 | Dld | dihydrolipoamide dehydrogenase [Source:MGI Symbol;Acc:MGI:107450] | 27620 | -0.699 | -0.5853 | Yes |
| 92 | Idh3g | isocitrate dehydrogenase 3 (NAD+), gamma [Source:MGI Symbol;Acc:MGI:1099463] | 27748 | -0.724 | -0.5835 | Yes |
| 93 | Suclg2 | succinate-Coenzyme A ligase, GDP-forming, beta subunit [Source:MGI Symbol;Acc:MGI:1306824] | 27814 | -0.733 | -0.5795 | Yes |
| 94 | Adh7 | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide [Source:MGI Symbol;Acc:MGI:87926] | 27856 | -0.739 | -0.5747 | Yes |
| 95 | Adipor2 | adiponectin receptor 2 [Source:MGI Symbol;Acc:MGI:93830] | 27879 | -0.745 | -0.5693 | Yes |
| 96 | Aadat | aminoadipate aminotransferase [Source:MGI Symbol;Acc:MGI:1345167] | 27904 | -0.748 | -0.5639 | Yes |
| 97 | Acads | acyl-Coenzyme A dehydrogenase, short chain [Source:MGI Symbol;Acc:MGI:87868] | 27917 | -0.750 | -0.5581 | Yes |
| 98 | D2hgdh | D-2-hydroxyglutarate dehydrogenase [Source:MGI Symbol;Acc:MGI:2138209] | 27986 | -0.769 | -0.5540 | Yes |
| 99 | Ugdh | UDP-glucose dehydrogenase [Source:MGI Symbol;Acc:MGI:1306785] | 28002 | -0.772 | -0.5481 | Yes |
| 100 | Hadh | hydroxyacyl-Coenzyme A dehydrogenase [Source:MGI Symbol;Acc:MGI:96009] | 28113 | -0.797 | -0.5451 | Yes |
| 101 | Retsat | retinol saturase (all trans retinol 13,14 reductase) [Source:MGI Symbol;Acc:MGI:1914692] | 28114 | -0.798 | -0.5385 | Yes |
| 102 | Aoc3 | amine oxidase, copper containing 3 [Source:MGI Symbol;Acc:MGI:1306797] | 28172 | -0.810 | -0.5336 | Yes |
| 103 | Sms-ps | spermine synthase, pseudogene [Source:MGI Symbol;Acc:MGI:3705601] | 28245 | -0.829 | -0.5291 | Yes |
| 104 | Acadm | acyl-Coenzyme A dehydrogenase, medium chain [Source:MGI Symbol;Acc:MGI:87867] | 28248 | -0.829 | -0.5224 | Yes |
| 105 | Blvra | biliverdin reductase A [Source:MGI Symbol;Acc:MGI:88170] | 28348 | -0.853 | -0.5185 | Yes |
| 106 | Acaa1a | acetyl-Coenzyme A acyltransferase 1A [Source:MGI Symbol;Acc:MGI:2148491] | 28500 | -0.888 | -0.5160 | Yes |
| 107 | Eno3 | enolase 3, beta muscle [Source:MGI Symbol;Acc:MGI:95395] | 28516 | -0.893 | -0.5092 | Yes |
| 108 | Eci2 | enoyl-Coenzyme A delta isomerase 2 [Source:MGI Symbol;Acc:MGI:1346064] | 28521 | -0.893 | -0.5019 | Yes |
| 109 | Psme1 | proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) [Source:MGI Symbol;Acc:MGI:1096367] | 28573 | -0.906 | -0.4961 | Yes |
| 110 | Alad | aminolevulinate, delta-, dehydratase [Source:MGI Symbol;Acc:MGI:96853] | 28677 | -0.937 | -0.4917 | Yes |
| 111 | Cd36 | CD36 molecule [Source:MGI Symbol;Acc:MGI:107899] | 28751 | -0.957 | -0.4862 | Yes |
| 112 | Bckdhb | branched chain ketoacid dehydrogenase E1, beta polypeptide [Source:MGI Symbol;Acc:MGI:88137] | 28761 | -0.958 | -0.4786 | Yes |
| 113 | Me1 | malic enzyme 1, NADP(+)-dependent, cytosolic [Source:MGI Symbol;Acc:MGI:97043] | 28808 | -0.972 | -0.4720 | Yes |
| 114 | Car2 | carbonic anhydrase 2 [Source:MGI Symbol;Acc:MGI:88269] | 28961 | -1.017 | -0.4685 | Yes |
| 115 | Cpt2 | carnitine palmitoyltransferase 2 [Source:MGI Symbol;Acc:MGI:109176] | 29065 | -1.048 | -0.4632 | Yes |
| 116 | Eci1 | enoyl-Coenzyme A delta isomerase 1 [Source:MGI Symbol;Acc:MGI:94871] | 29108 | -1.064 | -0.4558 | Yes |
| 117 | Aldh3a2 | aldehyde dehydrogenase family 3, subfamily A2 [Source:MGI Symbol;Acc:MGI:1353452] | 29334 | -1.134 | -0.4537 | Yes |
| 118 | Suclg1 | succinate-CoA ligase, GDP-forming, alpha subunit [Source:MGI Symbol;Acc:MGI:1927234] | 29356 | -1.142 | -0.4449 | Yes |
| 119 | Aldh9a1 | aldehyde dehydrogenase 9, subfamily A1 [Source:MGI Symbol;Acc:MGI:1861622] | 29361 | -1.145 | -0.4356 | Yes |
| 120 | Nthl1 | nth (endonuclease III)-like 1 (E.coli) [Source:MGI Symbol;Acc:MGI:1313275] | 29452 | -1.182 | -0.4288 | Yes |
| 121 | Adsl | adenylosuccinate lyase [Source:MGI Symbol;Acc:MGI:103202] | 29469 | -1.189 | -0.4195 | Yes |
| 122 | Cpox | coproporphyrinogen oxidase [Source:MGI Symbol;Acc:MGI:104841] | 29516 | -1.206 | -0.4110 | Yes |
| 123 | Fh1 | fumarate hydratase 1 [Source:MGI Symbol;Acc:MGI:95530] | 29519 | -1.209 | -0.4011 | Yes |
| 124 | Gcdh | glutaryl-Coenzyme A dehydrogenase [Source:MGI Symbol;Acc:MGI:104541] | 29607 | -1.243 | -0.3937 | Yes |
| 125 | Fasn | fatty acid synthase [Source:MGI Symbol;Acc:MGI:95485] | 29614 | -1.246 | -0.3836 | Yes |
| 126 | Mcee | methylmalonyl CoA epimerase [Source:MGI Symbol;Acc:MGI:1920974] | 29640 | -1.260 | -0.3740 | Yes |
| 127 | Acaa2 | acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase) [Source:MGI Symbol;Acc:MGI:1098623] | 29705 | -1.285 | -0.3655 | Yes |
| 128 | Mdh1 | malate dehydrogenase 1, NAD (soluble) [Source:MGI Symbol;Acc:MGI:97051] | 29716 | -1.290 | -0.3552 | Yes |
| 129 | Ostc | oligosaccharyltransferase complex subunit (non-catalytic) [Source:MGI Symbol;Acc:MGI:1913607] | 29816 | -1.331 | -0.3474 | Yes |
| 130 | Glul | glutamate-ammonia ligase (glutamine synthetase) [Source:MGI Symbol;Acc:MGI:95739] | 29848 | -1.346 | -0.3373 | Yes |
| 131 | Hibch | 3-hydroxyisobutyryl-Coenzyme A hydrolase [Source:MGI Symbol;Acc:MGI:1923792] | 29856 | -1.349 | -0.3264 | Yes |
| 132 | Uros | uroporphyrinogen III synthase [Source:MGI Symbol;Acc:MGI:98917] | 29924 | -1.385 | -0.3172 | Yes |
| 133 | Hsd17b10 | hydroxysteroid (17-beta) dehydrogenase 10 [Source:MGI Symbol;Acc:MGI:1333871] | 29972 | -1.413 | -0.3070 | Yes |
| 134 | Car6 | carbonic anhydrase 6 [Source:MGI Symbol;Acc:MGI:1333786] | 30090 | -1.483 | -0.2986 | Yes |
| 135 | Gstz1 | glutathione transferase zeta 1 (maleylacetoacetate isomerase) [Source:MGI Symbol;Acc:MGI:1341859] | 30139 | -1.515 | -0.2876 | Yes |
| 136 | Cbr3 | carbonyl reductase 3 [Source:MGI Symbol;Acc:MGI:1309992] | 30171 | -1.535 | -0.2759 | Yes |
| 137 | Hmgcl | 3-hydroxy-3-methylglutaryl-Coenzyme A lyase [Source:MGI Symbol;Acc:MGI:96158] | 30197 | -1.554 | -0.2639 | Yes |
| 138 | Ech1 | enoyl coenzyme A hydratase 1, peroxisomal [Source:MGI Symbol;Acc:MGI:1858208] | 30334 | -1.652 | -0.2547 | Yes |
| 139 | Grhpr | glyoxylate reductase/hydroxypyruvate reductase [Source:MGI Symbol;Acc:MGI:1923488] | 30381 | -1.688 | -0.2423 | Yes |
| 140 | Pts | 6-pyruvoyl-tetrahydropterin synthase [Source:MGI Symbol;Acc:MGI:1338783] | 30388 | -1.692 | -0.2285 | Yes |
| 141 | Bphl | biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) [Source:MGI Symbol;Acc:MGI:1915271] | 30399 | -1.699 | -0.2148 | Yes |
| 142 | Echs1 | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial [Source:MGI Symbol;Acc:MGI:2136460] | 30590 | -1.904 | -0.2052 | Yes |
| 143 | Slc22a5 | solute carrier family 22 (organic cation transporter), member 5 [Source:MGI Symbol;Acc:MGI:1329012] | 30597 | -1.914 | -0.1897 | Yes |
| 144 | Decr1 | 2,4-dienoyl CoA reductase 1, mitochondrial [Source:MGI Symbol;Acc:MGI:1914710] | 30605 | -1.924 | -0.1740 | Yes |
| 145 | Acadvl | acyl-Coenzyme A dehydrogenase, very long chain [Source:MGI Symbol;Acc:MGI:895149] | 30679 | -2.012 | -0.1598 | Yes |
| 146 | Reep6 | receptor accessory protein 6 [Source:MGI Symbol;Acc:MGI:1917585] | 30707 | -2.047 | -0.1438 | Yes |
| 147 | Mlycd | malonyl-CoA decarboxylase [Source:MGI Symbol;Acc:MGI:1928485] | 30803 | -2.170 | -0.1290 | Yes |
| 148 | Aldh1a1 | aldehyde dehydrogenase family 1, subfamily A1 [Source:MGI Symbol;Acc:MGI:1353450] | 30858 | -2.263 | -0.1120 | Yes |
| 149 | Acot2 | acyl-CoA thioesterase 2 [Source:MGI Symbol;Acc:MGI:2159605] | 30957 | -2.480 | -0.0947 | Yes |
| 150 | Cbr1 | carbonyl reductase 1 [Source:MGI Symbol;Acc:MGI:88284] | 30982 | -2.536 | -0.0746 | Yes |
| 151 | Aldh3a1 | aldehyde dehydrogenase family 3, subfamily A1 [Source:MGI Symbol;Acc:MGI:1353451] | 31130 | -3.120 | -0.0536 | Yes |
| 152 | Acsm3 | acyl-CoA synthetase medium-chain family member 3 [Source:MGI Symbol;Acc:MGI:99538] | 31154 | -3.363 | -0.0266 | Yes |
| 153 | G0s2 | G0/G1 switch gene 2 [Source:MGI Symbol;Acc:MGI:1316737] | 31168 | -3.556 | 0.0023 | Yes |
Table: GSEA details [plain text format]