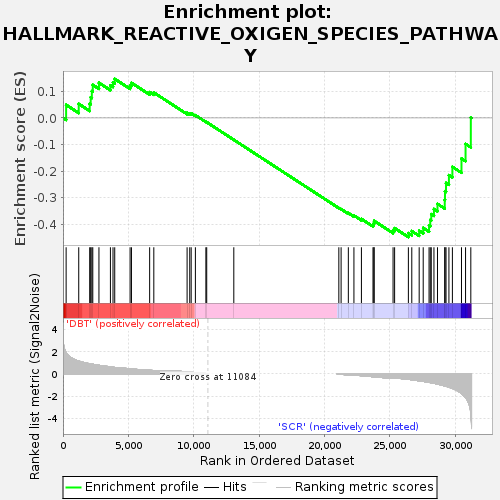
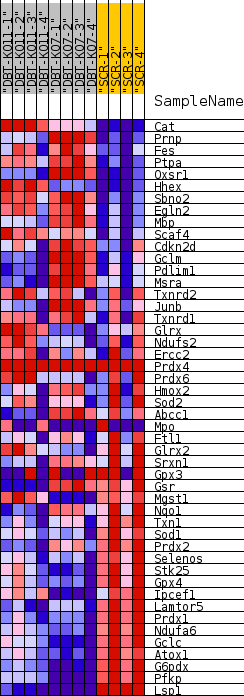
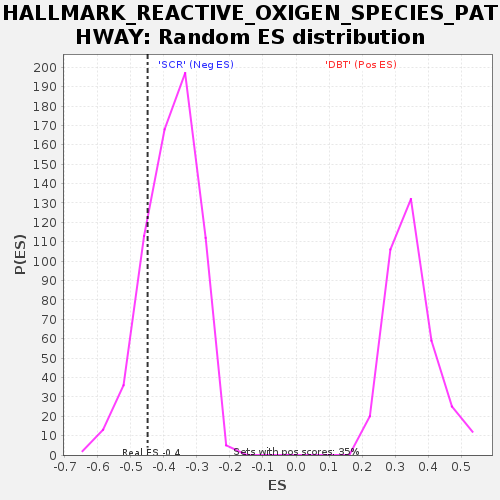
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | | 1 | Cat | catalase [Source:MGI Symbol;Acc:MGI:88271] | 239 | 1.883 | 0.0493 | No |
| 2 | Prnp | prion protein [Source:MGI Symbol;Acc:MGI:97769] | 1203 | 1.142 | 0.0530 | No |
| 3 | Fes | feline sarcoma oncogene [Source:MGI Symbol;Acc:MGI:95514] | 2041 | 0.898 | 0.0533 | No |
| 4 | Ptpa | protein phosphatase 2 protein activator [Source:MGI Symbol;Acc:MGI:1346006] | 2115 | 0.883 | 0.0777 | No |
| 5 | Oxsr1 | oxidative-stress responsive 1 [Source:MGI Symbol;Acc:MGI:1917378] | 2205 | 0.866 | 0.1010 | No |
| 6 | Hhex | hematopoietically expressed homeobox [Source:MGI Symbol;Acc:MGI:96086] | 2277 | 0.852 | 0.1245 | No |
| 7 | Sbno2 | strawberry notch 2 [Source:MGI Symbol;Acc:MGI:2448490] | 2748 | 0.759 | 0.1324 | No |
| 8 | Egln2 | egl-9 family hypoxia-inducible factor 2 [Source:MGI Symbol;Acc:MGI:1932287] | 3625 | 0.613 | 0.1228 | No |
| 9 | Mbp | myelin basic protein [Source:MGI Symbol;Acc:MGI:96925] | 3823 | 0.582 | 0.1341 | No |
| 10 | Scaf4 | SR-related CTD-associated factor 4 [Source:MGI Symbol;Acc:MGI:2146350] | 3955 | 0.562 | 0.1470 | No |
| 11 | Cdkn2d | cyclin dependent kinase inhibitor 2D [Source:MGI Symbol;Acc:MGI:105387] | 5129 | 0.442 | 0.1227 | No |
| 12 | Gclm | glutamate-cysteine ligase, modifier subunit [Source:MGI Symbol;Acc:MGI:104995] | 5234 | 0.425 | 0.1323 | No |
| 13 | Pdlim1 | PDZ and LIM domain 1 (elfin) [Source:MGI Symbol;Acc:MGI:1860611] | 6625 | 0.313 | 0.0972 | No |
| 14 | Msra | methionine sulfoxide reductase A [Source:MGI Symbol;Acc:MGI:106916] | 6943 | 0.285 | 0.0956 | No |
| 15 | Txnrd2 | thioredoxin reductase 2 [Source:MGI Symbol;Acc:MGI:1347023] | 9488 | 0.167 | 0.0191 | No |
| 16 | Junb | jun B proto-oncogene [Source:MGI Symbol;Acc:MGI:96647] | 9691 | 0.144 | 0.0170 | No |
| 17 | Txnrd1 | thioredoxin reductase 1 [Source:MGI Symbol;Acc:MGI:1354175] | 9800 | 0.130 | 0.0175 | No |
| 18 | Glrx | glutaredoxin [Source:MGI Symbol;Acc:MGI:2135625] | 10122 | 0.103 | 0.0103 | No |
| 19 | Ndufs2 | NADH:ubiquinone oxidoreductase core subunit S2 [Source:MGI Symbol;Acc:MGI:2385112] | 10924 | 0.026 | -0.0146 | No |
| 20 | Ercc2 | excision repair cross-complementing rodent repair deficiency, complementation group 2 [Source:MGI Symbol;Acc:MGI:95413] | 10988 | 0.017 | -0.0161 | No |
| 21 | Prdx4 | peroxiredoxin 4 [Source:MGI Symbol;Acc:MGI:1859815] | 13059 | 0.000 | -0.0825 | No |
| 22 | Prdx6 | peroxiredoxin 6 [Source:MGI Symbol;Acc:MGI:894320] | 21089 | -0.023 | -0.3392 | No |
| 23 | Hmox2 | heme oxygenase 2 [Source:MGI Symbol;Acc:MGI:109373] | 21263 | -0.044 | -0.3434 | No |
| 24 | Sod2 | superoxide dismutase 2, mitochondrial [Source:MGI Symbol;Acc:MGI:98352] | 21816 | -0.099 | -0.3581 | No |
| 25 | Abcc1 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 [Source:MGI Symbol;Acc:MGI:102676] | 22243 | -0.145 | -0.3674 | No |
| 26 | Mpo | myeloperoxidase [Source:MGI Symbol;Acc:MGI:97137] | 22816 | -0.171 | -0.3805 | No |
| 27 | Ftl1 | ferritin light polypeptide 1 [Source:MGI Symbol;Acc:MGI:95589] | 23701 | -0.258 | -0.4011 | No |
| 28 | Glrx2 | glutaredoxin 2 (thioltransferase) [Source:MGI Symbol;Acc:MGI:1916617] | 23773 | -0.264 | -0.3954 | No |
| 29 | Srxn1 | sulfiredoxin 1 homolog (S. cerevisiae) [Source:MGI Symbol;Acc:MGI:104971] | 23786 | -0.266 | -0.3877 | No |
| 30 | Gpx3 | glutathione peroxidase 3 [Source:MGI Symbol;Acc:MGI:105102] | 25235 | -0.369 | -0.4230 | No |
| 31 | Gsr | glutathione reductase [Source:MGI Symbol;Acc:MGI:95804] | 25354 | -0.383 | -0.4152 | No |
| 32 | Mgst1 | microsomal glutathione S-transferase 1 [Source:MGI Symbol;Acc:MGI:1913850] | 26406 | -0.496 | -0.4339 | Yes |
| 33 | Nqo1 | NAD(P)H dehydrogenase, quinone 1 [Source:MGI Symbol;Acc:MGI:103187] | 26667 | -0.534 | -0.4261 | Yes |
| 34 | Txn1 | thioredoxin 1 [Source:MGI Symbol;Acc:MGI:98874] | 27232 | -0.634 | -0.4250 | Yes |
| 35 | Sod1 | superoxide dismutase 1, soluble [Source:MGI Symbol;Acc:MGI:98351] | 27540 | -0.679 | -0.4143 | Yes |
| 36 | Prdx2 | peroxiredoxin 2 [Source:MGI Symbol;Acc:MGI:109486] | 27987 | -0.769 | -0.4053 | Yes |
| 37 | Selenos | selenoprotein S [Source:MGI Symbol;Acc:MGI:95994] | 28078 | -0.789 | -0.3843 | Yes |
| 38 | Stk25 | serine/threonine kinase 25 (yeast) [Source:MGI Symbol;Acc:MGI:1891699] | 28156 | -0.806 | -0.3624 | Yes |
| 39 | Gpx4 | glutathione peroxidase 4 [Source:MGI Symbol;Acc:MGI:104767] | 28361 | -0.856 | -0.3431 | Yes |
| 40 | Ipcef1 | interaction protein for cytohesin exchange factors 1 [Source:MGI Symbol;Acc:MGI:2444159] | 28632 | -0.926 | -0.3237 | Yes |
| 41 | Lamtor5 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 [Source:MGI Symbol;Acc:MGI:1915826] | 29187 | -1.086 | -0.3087 | Yes |
| 42 | Prdx1 | peroxiredoxin 1 [Source:MGI Symbol;Acc:MGI:99523] | 29206 | -1.092 | -0.2762 | Yes |
| 43 | Ndufa6 | NADH:ubiquinone oxidoreductase subunit A6 [Source:MGI Symbol;Acc:MGI:1914380] | 29285 | -1.118 | -0.2449 | Yes |
| 44 | Gclc | glutamate-cysteine ligase, catalytic subunit [Source:MGI Symbol;Acc:MGI:104990] | 29506 | -1.204 | -0.2155 | Yes |
| 45 | Atox1 | antioxidant 1 copper chaperone [Source:MGI Symbol;Acc:MGI:1333855] | 29771 | -1.315 | -0.1842 | Yes |
| 46 | G6pdx | glucose-6-phosphate dehydrogenase X-linked [Source:MGI Symbol;Acc:MGI:105979] | 30467 | -1.772 | -0.1529 | Yes |
| 47 | Pfkp | phosphofructokinase, platelet [Source:MGI Symbol;Acc:MGI:1891833] | 30778 | -2.141 | -0.0981 | Yes |
| 48 | Lsp1 | lymphocyte specific 1 [Source:MGI Symbol;Acc:MGI:96832] | 31180 | -3.729 | 0.0019 | Yes |
Table: GSEA details [plain text format]